Giulio Regeni. Until There Is Justice - ChannelDraw
An artistic journey at the heart of an open wound
10 years ago today, Italian PhD student Giulio Regeni was found murdered after being kidnapped and tortured by Egyptian security services. Egypt has never cooperated with Italian prosecutors to identify those responsible
www.channeldraw.org/2026/01/24/g...
02.02.2026 06:18 — 👍 0 🔁 0 💬 0 📌 0

Growth-coupled microbial biosynthesis of the animal pigment xanthommatin — Led by Leah Bushin, featuring M. Gracia Alvan, @danielvolke.bsky.social @oscarpuiggene.bsky.social in a fantastic collaboration w/Brad Moore @labnikel.bsky.social @natbiotech.nature.com 🦑
www.nature.com/articles/s41...
03.11.2025 12:49 — 👍 33 🔁 15 💬 2 📌 1
Redirecting
🔥🔥🔥 Friday Evening, Paper Out, Feierabend 🔥🔥🔥
When evolution and enzyme engineering team up, they can convince microbes to assimilate sustainable carbon substrates preparing them for a life in a circular bioeconomy.
doi.org/10.1016/j.ym...
24.10.2025 17:00 — 👍 7 🔁 6 💬 3 📌 0
Thanks! First challenge is to expand the substrate acceptance range in the cell factory
22.09.2025 14:31 — 👍 1 🔁 0 💬 1 📌 0
Please repost!
Postdoc in adaptive laboratory evolution and C1 synthetic metabolism!
Location: DTU Biosustain
🦠🧬🔬
Starting in 01/2026!
Fell free to reach out if you have questions!
efzu.fa.em2.oraclecloud.com/hcmUI/Candid...
12.09.2025 11:43 — 👍 8 🔁 16 💬 1 📌 0
🧪 We're hiring a #Postdoc in Bioprocess Engineering!
Excited about scaling CO₂-based biomanufacturing from bench to pilot scale?
Join our group working on converting CO₂ + renewable electricity → polymers 💡🌱 👇
10.07.2025 14:26 — 👍 1 🔁 2 💬 1 📌 0
#ME16
This is quite subjective , but imo
@helenasm.bsky.social just gave the most impressive talk of the conference so far and she is "only" in the third year of her PhD.
If you are interested in synthetic pathway engineering or C1 fixation you should check out her research.
16.06.2025 13:45 — 👍 5 🔁 2 💬 1 📌 0

🎉 Excited to share that I’ve received an Emerging Investigator Grant from the Novo Nordisk Foundation! I’ll start my research group in Jan 2026 to develop universal cell factories for P2X & CO₂-based biomanufacturing 🌱🔬
👉 researchleaderprogramme.com/recipients/e...
03.06.2025 13:56 — 👍 15 🔁 4 💬 1 📌 1
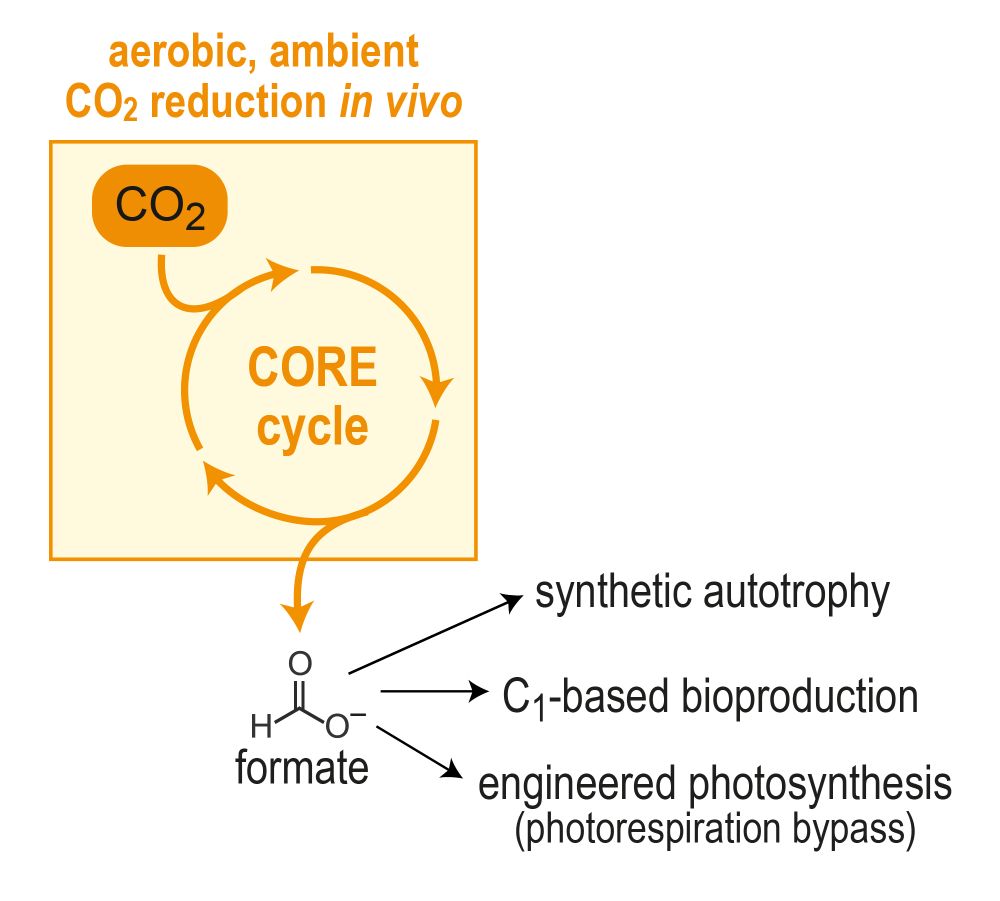
Excited to share a main project from my PhD, out now in
@naturecomms.bsky.social! 📝
We've designed and brought to life the “CORE cycle” – a new-to-nature pathway that provides a novel route for biological CO2 capture 🦠🌱
nature.com/articles/s41...
Take a look! Thread below... 🧵
04.04.2025 10:03 — 👍 39 🔁 17 💬 4 📌 4
Redirecting
Excited to see our SIBR work out in Trends in Biotechnology :)
doi.org/10.1016/j.ti...
This is the first study I have coordinated from start to finish, and it brings me immense satisfaction to see it published.
For a brilliant description of the tech check this: www.linkedin.com/feed/update/...
14.03.2025 10:37 — 👍 5 🔁 1 💬 0 📌 0
We had great support from our labs in developing this toolkit. Thanks to @pabnik.bsky.social, @nicoc-micsynmet.bsky.social, Sjoerd, Luc, Angela, Chase, Harrison, Raymond, John, and Wei!
The plasmids will be available on Addgene. Feel free to reach out in case you want to try them!
14.03.2025 11:16 — 👍 2 🔁 0 💬 0 📌 0

These results demonstrates that a new state-of-the-art is available for making gene deletions in this promising bug.
With a turnaround time that is 50% shorter than the current systems available, we believe that SIBR-Cas9 and SIBR2.0-Cas12a will help using C. necator in new P2X applications! 8/n
14.03.2025 11:16 — 👍 1 🔁 0 💬 1 📌 0

SIBR2.0 was then functional and we moved testing it for Cas12a. The new architectures revealed to be tight, allowing precise and controllable induction of cas12a as well, with similar efficiencies to the ones observed for Cas9! 7/n
14.03.2025 11:16 — 👍 0 🔁 0 💬 1 📌 0

Here is where SIBR2.0 comes into place. Simona and Costas developed a script for strategically moving the intron position within the CDS to get rid of such hidden translation initiation starts. This was tested first in E. coli using GFP as readout. 6/n
14.03.2025 11:16 — 👍 0 🔁 0 💬 1 📌 0

Then, we moved at testing if SIBR would work also on Cas12a. To our surprise, induction during the targeting assay revealed a leaky cas12a expression despite the presence of the intron with the STOP codon. This was because of a hidden translation start site that required some adjustments. 5/n
14.03.2025 11:16 — 👍 0 🔁 0 💬 1 📌 0

We started by testing the SIBR setup on Cas9. This was because we already had evidence that it worked before, although poorly www.sciencedirect.com/science/arti...
Targeting worked fine & by combining SIBR w/ Cas9, we achieved high editing efficiencies of >80% in two loci! Great start! 4/n
14.03.2025 11:16 — 👍 0 🔁 0 💬 1 📌 0

Streamlined CRISPR genome engineering in wild-type bacteria using SIBR-Cas
Abstract. CRISPR-Cas is a powerful tool for genome editing in bacteria. However, its efficacy is dependent on host factors (such as DNA repair pathways) an
Teaming up with Simona and Costas, we decided to apply the SIBR technology that Costas developed during his PhD to "pause" Cas counter-selection and allow endogenous homologous recombination (HR) to happen first. Our suspect was that C. necator is also poor at HR academic.oup.com/nar/article/... 3/n
14.03.2025 11:16 — 👍 0 🔁 0 💬 1 📌 0
C. necator holds potential for the conversion and valorization of CO2 for its ability to grow on this substrate using power-to-X feedstocks (like hydrogen or formate). However, making gene deletions in this organism has always been a pain due to its limited gene deletion toolkit 2/n
14.03.2025 11:16 — 👍 0 🔁 0 💬 1 📌 0
Check out Sebastians crazy effort to engineer E. coli to grow on formate via the synthetic Serine-Threonine-Cycle. In contrast to the rGlyp the STC works even at ambient CO2!🔥
06.03.2025 12:07 — 👍 4 🔁 2 💬 0 📌 0

PhD aminosugar metabolism
The Faculty of Science / the Institute for Biology is looking for a :PhD Candidate , Aminosugar metabolism and control of growth and development in bacteriaVacancy number: 15487 PhD project descriptio...
Only one week left to apply for this exciting PhD position, supervised by @gillesvanwezel.bsky.social and me!
Please apply if you are interested in microbial biochemistry and have a solid background in enzymes, metabolic pathways, and bacterial genetics.
www.universiteitleiden.nl/en/vacancies...
06.03.2025 17:08 — 👍 2 🔁 5 💬 0 📌 0
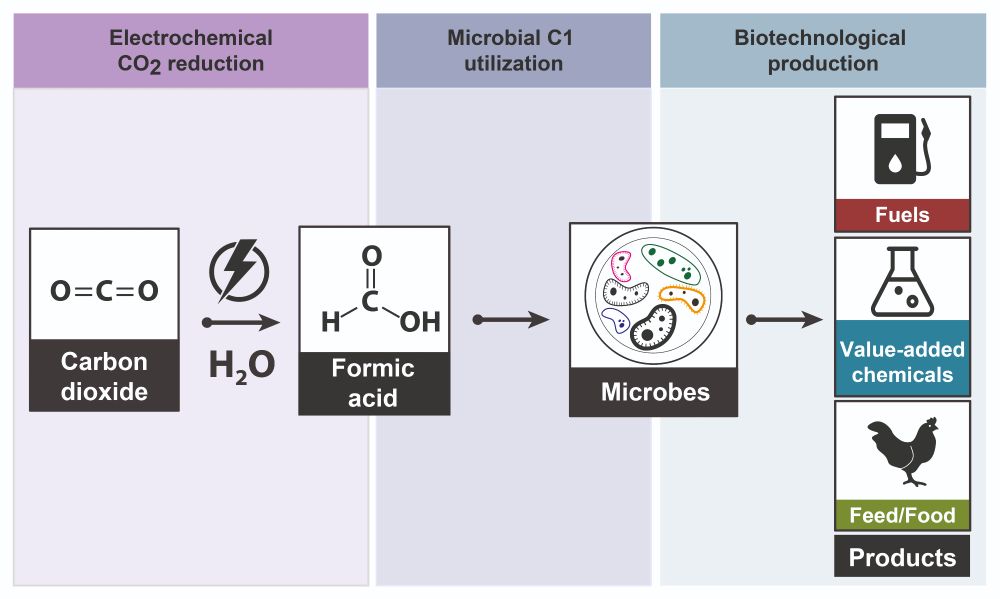
Interested in Sustainability, Metabolic Engineering and Evolution?
Check out how we convinced E. coli to grow on the CO2-derived substrate formic acid using the synthetic Serine Threonine Cycle! ♻️🌱🦠
OPEN ACCESS publication to be found here:
🚨 doi.org/10.1016/j.ym... 🚨
#MEvoSky #microsky
(1/🧵)
06.03.2025 10:11 — 👍 37 🔁 14 💬 2 📌 2
Prof. of Microbial Biotechnology at BOKU University Vienna, Austria
metabolic engineering • synthetic biology • C1 biotechnology • yeast • home baking • art & science
PhD student rewiring E. coli for synthetic C1 assimilation
Assistant Professor at the University of Zurich.
Alumni @schuhlab and @IsabelleVernos.
Cycling biologist.
Lab website: www.reproendo.uzh.ch/en/CavazzaLab.html
Scientific Coordinator of the Neuland project at IPK Gatersleben. Agronomist, PhD in plant biology. Soil ecology, plant–microbe interactions, and a soft spot for data. Sometimes acting on stages.
Contact: bluesky@rosario.mozmail.com
Group leader at VU LSC-EMBL PI for Genome Editing Technologies, Vilnius, Lithuania
Genome engineer
CRISPR-Cas
DNA replication stall and repair
Professor DTU Biosustain · Proud PI @labnikel.bsky.social · SynBio · Metabolic Engineering · @novo-nordisk.bsky.social Ascending Investigator & Distinguished Innovator · He/him🏳️🌈
Microbial engineering enthusiast. Synthetic Biology. Sustainable Biotechnology. Closing the carbon loop.
https://orcid.org/0000-0001-9065-6764
https://dixonlab.org
Scientist at the CNB-CSIC Madrid
SynBio enthusiast 🧬🦠
Microbial engineering | Synthetic metabolism | Biosensors
Berlin 🇩🇪 | Madrid 🇪🇸 | Groningen 🇳🇱
In love with single molecules 🔬 moving inside tiny things 🦠 and big dumb monsters 🦖 wreaking havoc on the screen 🎬
Postdoctoral researcher @ Laboratory of Microbiology (WUR)
Also on Letterboxd -> https://boxd.it/5Z17P
Synthetic Biology & Synthetic Genomics @ Imperial College London and the Sanger Institute. Bilingual in English and DNA. Views are either my own or my microbes'
PhD student at DTU Biosustain. Working on fungal GPCRs and more. - 1/3 of Biologi per la scienza
Thomas Willing Early Career Assoc. Professor, U. Delaware | #Synbio with newer-to-nature building blocks |
Formerly @Texas @MIT @HMS
www.kunjapurlab.org
Synthetic biology designer
Synthetic Biologist, Protein and Genome Engineer, Microbe and Fungi Fan
www.billerbecklab.com
#SyntheticBiology | #Genome Design & Genetic Code Engineering | Protein, RNA, DNA #DirectedEvolution beyond the boundaries of Nature |
Research Associate at Harvard | #NIH #NIBIB K99/R00
https://engineeringbio.science
Assistant Professor at the Institute of Biology Leiden | Microbial biochemistry, synthetic biology, environmental sustainability
Professor of Microbiome Ecology at Institute of Biology, Leiden University
Bacillus subtilis, biofilms, plant microbiome, bacteria-fungi interactions and circadian clock within @microclockerc.bsky.social #ERCSyG
https://linktr.ee/atkovacs
co-founder and CEO at Ginkgo Bioworks
Former Chair of National Security Commission on Emerging Biotechnology