

I had an amazing time yesterday visiting @vdlorenzo.bsky.social and hearing about the many exciting projects underway in his lab. Our evening stroll through the heart of Madrid and dinner of tapas made for an unforgettable experience
05.11.2025 17:34 — 👍 6 🔁 0 💬 0 📌 1
It's an honor to visit @vdlorenzo.bsky.social and colleagues at El Centro Nacional de Biotecnología in beautiful Madrid, España. Me alegra estar aquí
04.11.2025 08:09 — 👍 5 🔁 0 💬 0 📌 0


My students have become very good at cloning. Now I can be in two continents at once this fall
13.08.2025 12:58 — 👍 3 🔁 0 💬 0 📌 0
Excited to announce our new methods paper in Methods in Enzymology!
We share our pipeline for building high-diversity DNA libraries & exploring protein stability in E. coli.
Free access until Sept 4: authors.elsevier.com/a/1lRkeHRzCX...
Updated, companion preprint: www.biorxiv.org/content/10.1...
17.07.2025 14:24 — 👍 5 🔁 3 💬 0 📌 0
Exciting to see this collaborative work about modeling soil microbiomes from the Kuehn, Mani, and Tikhonov labs published today
Also delighted to have lead author Kiseok Lee start his postdoc in our lab
Check out the paper below!
17.07.2025 00:22 — 👍 4 🔁 2 💬 0 📌 0
Looking forward to the amazing program & making new connections at the 17th International Symposium on Biocatalysis and Biotransformations (Biotrans 2025) starting today in Basel
I'm honored to present our work on Tuesday at 15:15 in the non-canonical amino acids session
#biocatalysis
29.06.2025 08:08 — 👍 5 🔁 0 💬 1 📌 0

Wrapping up a fantastic #ME16 #Copenhagen — a success by all measures! Honored to co-chair the meeting w/I. Borodina, M. Chang, A. Mirończuk, C. Vickers, V. Šťovíček, Y. Zhou, M. Köpke & M. Herrgård & to work w/an outstanding team of colleagues, engaging communities & topics all around the world
22.06.2025 19:46 — 👍 12 🔁 1 💬 0 📌 0
Congrats on completing your PhD @kiseokmicro.bsky.social!
And welcome to our lab - very excited to work with you
16.06.2025 08:52 — 👍 4 🔁 0 💬 0 📌 0
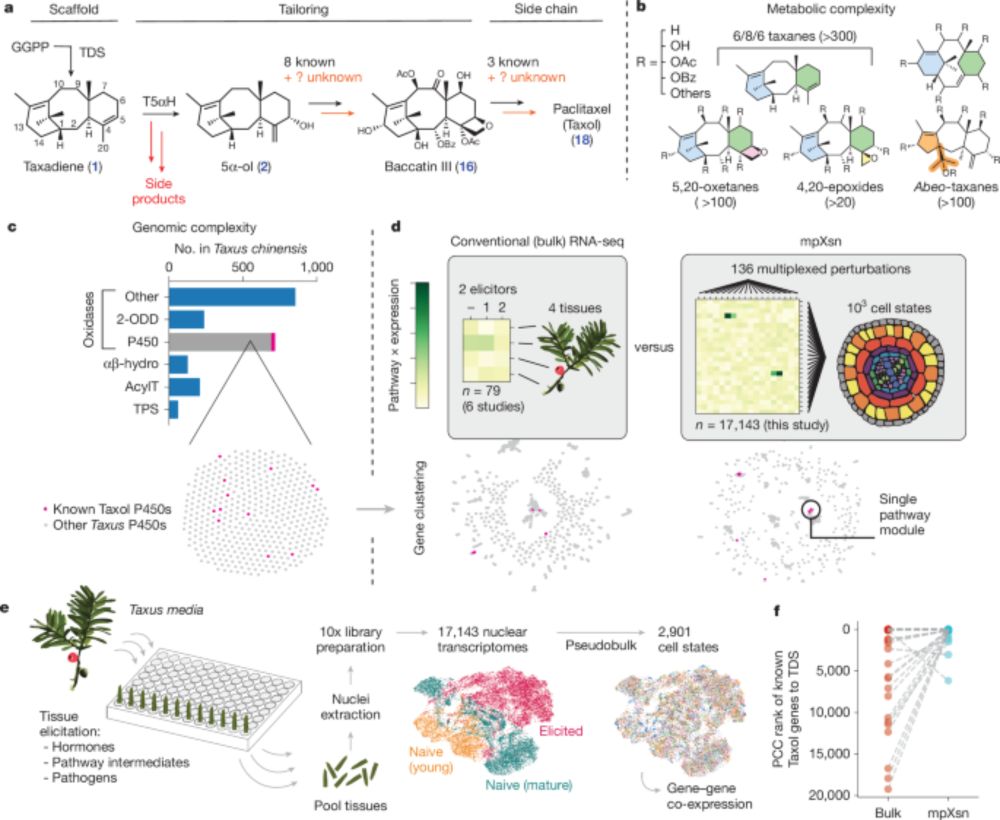
Discovery of FoTO1 and Taxol genes enables biosynthesis of baccatin III - Nature
An approach that combines single-nucleus RNA sequencing and multiplexed perturbation identifies genes that enable the biosynthesis of direct precursors of the anti-cancer drug Taxol, whose curren...
Today in Nature we report a systematic strategy to activate & identify gene sets in plants. We identify 8 new genes from the yew tree's Taxol biosynthetic pathway, enabling us to engineer tobacco with 17- & 20-gene pathways to Taxol precursors baccatin III & deBz-deoxy-Taxol #SingleCell #secmet 🧬🧪🌾
11.06.2025 19:49 — 👍 115 🔁 43 💬 3 📌 6
It is a real honor and delight to be hosted by @pabnik.bsky.social at DTU Biosustain!
Looking forward to meeting folks & sharing info/ideas
09.06.2025 10:27 — 👍 4 🔁 0 💬 0 📌 0
I don't take tenure for granted anymore as universities are under unprecedented attack
I'm also looking forward to sabbatical. I'll start as an Otto Mønsted Visiting Professor at Denmark Technical University next week and look forward to connecting more with my European colleagues
(3/3)
23.05.2025 19:37 — 👍 7 🔁 0 💬 0 📌 0


With the state of US scientific funding it seems like an inappropriate time to celebrate anything
But in the last week thanks to years of dedication from my students we were recognized through the Roberta Colman and Gerard Mangone awards, & tenure. Thank you to those who made it possible (1/3)
23.05.2025 19:31 — 👍 10 🔁 0 💬 1 📌 0
If you're curious how the first 5 amino acids of newly generated protein N-termini can affect protein stability in E. coli, then check out our latest preprint
We find new patterns of exceptions to the usual N-end rule
Kudos to Sunny Sen & co-authors
#synbio #protein_degradation
23.05.2025 19:05 — 👍 24 🔁 14 💬 0 📌 0
Thank you, Abhishek! Very glad you liked it
01.05.2025 19:21 — 👍 1 🔁 0 💬 0 📌 0
Thanks, Pat!!
Hope you've been doing well.
All three of the contexts you mentioned are under investigation by some combination of us, us and collaborators, or folks we have shared synthetic auxotrophs with
01.05.2025 15:07 — 👍 1 🔁 0 💬 1 📌 0
A more accurate statement than my original post:
It seems there may be few known pairs of microbial species, outside of obligate intracellular symbionts/parasites, where one relies exclusively on another.
Using a blend of synthetic and chemical biology, we designed such an interaction
01.05.2025 14:27 — 👍 6 🔁 1 💬 0 📌 1
Ah, thanks for sharing this example. "Free living" may be a bit of a misnomer, but you correctly interpreted that I wanted to exclude the category of obligate intracellular symbionts/parasites
01.05.2025 13:53 — 👍 0 🔁 0 💬 0 📌 0
This work represents a first step towards developing safeguards that could contain and sustain engineered microbes in a specific environment
That could be useful for addressing myriad challenges in planetary health
Work led by Amanda Forti, who is tenacious, meticulous, & entering the job market
01.05.2025 10:31 — 👍 2 🔁 2 💬 1 📌 0
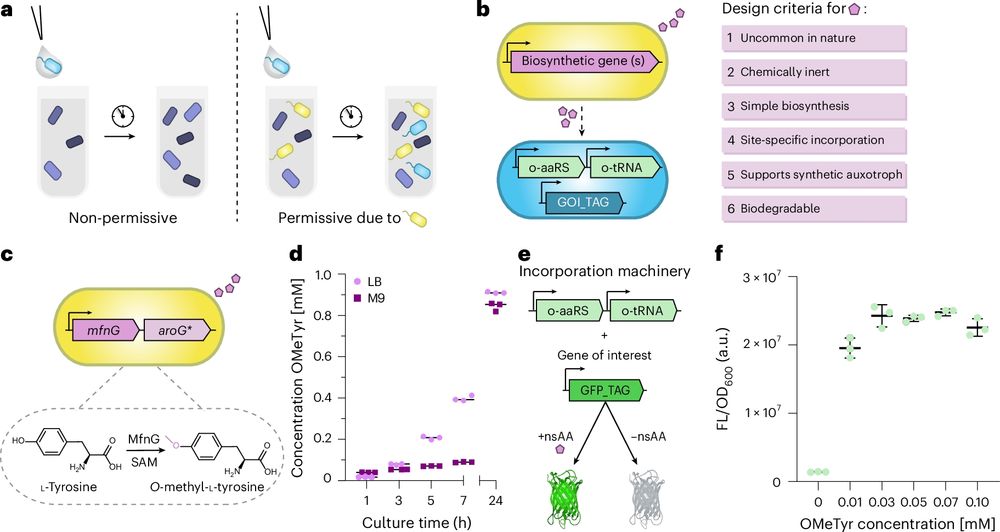
Engineered orthogonal and obligate bacterial commensalism mediated by a non-standard amino acid
Nature Microbiology - A genetically engineered microbial symbiosis based on synthetic auxotrophy is created, with potential implications as a biocontainment strategy.
It seems there are no free-living organisms known to rely exclusively on one other organism for their survival
Until perhaps now
Check out our latest publication in @natmicrobiol.nature.com to see how we designed an exclusive microbial reliance using GCE
rdcu.be/ekepI
1/2
01.05.2025 10:30 — 👍 43 🔁 18 💬 3 📌 2
Thanks, Tae Seok!
28.03.2025 04:12 — 👍 1 🔁 0 💬 0 📌 0
This move could singlehandedly dismantle American leadership in science and technology innovation.
It goes into effect on Monday. How will universities adjust their budgets to support critical ongoing research?
08.02.2025 01:52 — 👍 3 🔁 1 💬 0 📌 0
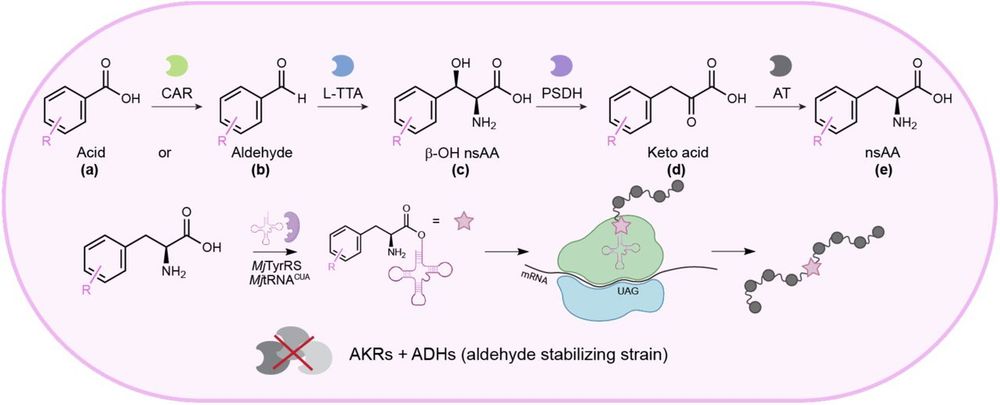
Overview of the engineered biosynthesis and incorporation platform. A single strain was designed to express two to four enzymes for the biosynthesis of phenylalanine derivates from supplemented acid or aldehyde precursors. The biosynthesized nsAA was then incorporated into a target protein using amber stop codon suppression and an orthogonal translation system. Source: https://www.biorxiv.org/content/10.1101/2024.12.17.628963v1.full

Biological materials are built with a limited number of building blocks based on polysaccharides, proteins, and minerals. A diversity of structures leads to a diversity of functions in tooth, bone, artery wall, tendon, spider web, beak, feather, wool, fingernail, tree, cotton, beetle carapace, lobster shell, snail shell, mussel shell, and the skeleton of the glass sponge (clockwise from top). Primary source: https://www.science.org/doi/10.1126/science.aat8297
2025-01-12 Issue of #engineeredlivingmaterials feed
biomed.news/bims-enlima/...
h/t @biomednews.bsky.social
includes @kunjapur.bsky.social lab's preprint @biorxivpreprint.bsky.social on combined biosynthesis and site-specific incorporation of phenylalanine derivatives in engineered bacteria
17.01.2025 12:32 — 👍 4 🔁 3 💬 0 📌 0
Biologist. Working on novel synthetic biology chassis, dynamic input sensing, non-canonical synthetic gene circuits and evolution. Currently at CNB-CSIC.
Chemistry postdoc, TU Delft
Interdisciplinary organization led by Dr. Steven Benner combining chemistry, synthetic biology & astrobiology, translating basic research into medical and commercial technologies. Visit our website at https://ffame.org/.
Virginie Hamel & Paul Guichard Lab at University of Geneva
#cryoEM/ET❄️ and #UExM ⚗️ #ExpansionMicroscopy #TeamTomo
Genève, Suisse 🇨🇭
https://mocel.unige.ch/research-groups/guichard-hamel/overv
Associate Professor for Molecular Evolution at the University of Groningen. My group makes biomolecules to order by letting millions of them compete until a winner is crowned :) Sometimes it even works!
Interdisciplinary group at the University of Groningen @rug.nl working on Artificial Enzymes, Bioorthogonal Catalysis and Chemistry in Living Cells. Posts by group members.
https://sites.google.com/rug.nl/roelfesgroup/
MSCA fellow at @crg.eu w/M. Dias and @jonnyfrazer.bsky.social
> Biological Physics | Proteins | Comp Bio | ML
https://scholar.google.com/citations?user=n55NtEsAAAAJ&hl=en
PhD at the University of Adelaide | Synbio, CryoEM, X-ray crystallography, Biocatalysts |
Synthetic biology & biological engineering. Recent PhD graduate from UD CBE. Music, cooking, and lifting too.
NYC; Lab of Environmental Microbiology @rockefelleruniv.bsky.social
Deeply curious about Life, the Universe and Everything
Evolution · SynBio · Gene Regulation · Microbes
Group Leader @EPFL.bsky.social ELISIR Scholar - Systems and Synthetic Evolutionary Biology Lab
PostDoc @Wagner Lab UZH
🇧🇷🌈 he/him
The backend of biotech. AI, datacenters, and the systems powering the future of life sciences.
PhD student in biology at TU Darmstadt.
jeremie.marlhens.bio
Researching RNA-based logic gates in cell-free systems.
Exploring the intersection of engineering and biology.
Professor in the Evolution of Microbial Symbiosis and Deputy Head of Department of Biology, University of Copenhagen. Views are my own. He/him 🏳️🌈
https://www.linkedin.com/in/michael-poulsen-5804b926/
https://www.socialsymbioticevolution.com/
Protein biochemistry and computational biophysics. Postdoc at the Linderstrøm Lang Centre for Protein Science, University of Copenhagen.
Principal Investigator | Northwestern University Microbiology-Immunology | #RNA #ribosome #antibiotic resistance #AMR #MRSA | hails from 🇲🇾 by way of 🇹🇼 |Views are my own. 🧪 🧬🔬💉
(he/him) Former PhD student / Postdoc @ Korbel group, EMBL.
Protein ML person, mathematics & science enthusiast.
developer of salad
preprint: https://www.biorxiv.org/content/10.1101/2025.01.31.635780v1
code: https://github.com/mjendrusch/salad
Assoc prof @UDelaware | social neuroscientist interested "us vs them" and "human vs AI" | Mexican, Indigenous, Japanese First Gen | #UDPBS | @UDPOSCIR | 🧠
https://www.ifsnlab.org/
Protein and coffee lover, father of two, professor of biophysics and sudo scientist at the Linderstrøm-Lang Centre for Protein Science, University of Copenhagen 🇩🇰





















