
🔗 Discover more: bit.ly/3LUzM2U
#EMBO #GenomeEditing #CRISPR #SyntheticBiology #Microbiology #Biotechnology #Vilnius #LifeSciences
@copabio.bsky.social
Group leader at VU LSC-EMBL PI for Genome Editing Technologies, Vilnius, Lithuania Genome engineer CRISPR-Cas DNA replication stall and repair

🔗 Discover more: bit.ly/3LUzM2U
#EMBO #GenomeEditing #CRISPR #SyntheticBiology #Microbiology #Biotechnology #Vilnius #LifeSciences
This work will drive new genome editing technologies and uncover fundamental DNA repair mechanisms, with impact across microbiome research, synthetic biology, industrial biotechnology, and healthcare.
20.01.2026 15:30 — 👍 2 🔁 0 💬 1 📌 0🧬 My project, “Pause-Repair-Edit,” aims to move beyond current CRISPR tools by developing safer genome editing approaches that pause DNA replication to activate natural repair pathways—opening genome editing to many microorganisms that are currently hard to engineer.
20.01.2026 15:30 — 👍 0 🔁 0 💬 1 📌 0🚀 Excited to share that I’ve been awarded an EMBO Installation Grant to establish my research group at the EMBL Partnership Institute, Vilnius University Life Sciences Center! The grant provides €50,000/year for 5 years to build an internationally competitive lab and advance genome editing research.
20.01.2026 15:30 — 👍 8 🔁 1 💬 1 📌 0
Our paper is out! Hiding in plain sight among Cas12a nucleases, Cas12a3 cleaves not its RNA target but the 3′ ends of tRNA. Huge thanks to all who made this possible, especially the Beisel lab, Biao & Dirk for the structure, & @sebastianglatt.bsky.social for all things tRNA. doi.org/10.1038/s415...
07.01.2026 16:15 — 👍 34 🔁 20 💬 1 📌 0
Openness means more than access.
We’re pleased to announce that we now publish Replication Studies, helping make science more reliable, transparent, and trustworthy.
buff.ly/Gebypvq
Thank you Jens
07.09.2025 21:24 — 👍 1 🔁 0 💬 0 📌 0Thank you Nicholas 😊
05.09.2025 09:40 — 👍 1 🔁 0 💬 0 📌 0Amazing Martin!! All the best with the new lab and the amazing research you do!!!
05.09.2025 09:38 — 👍 1 🔁 0 💬 0 📌 0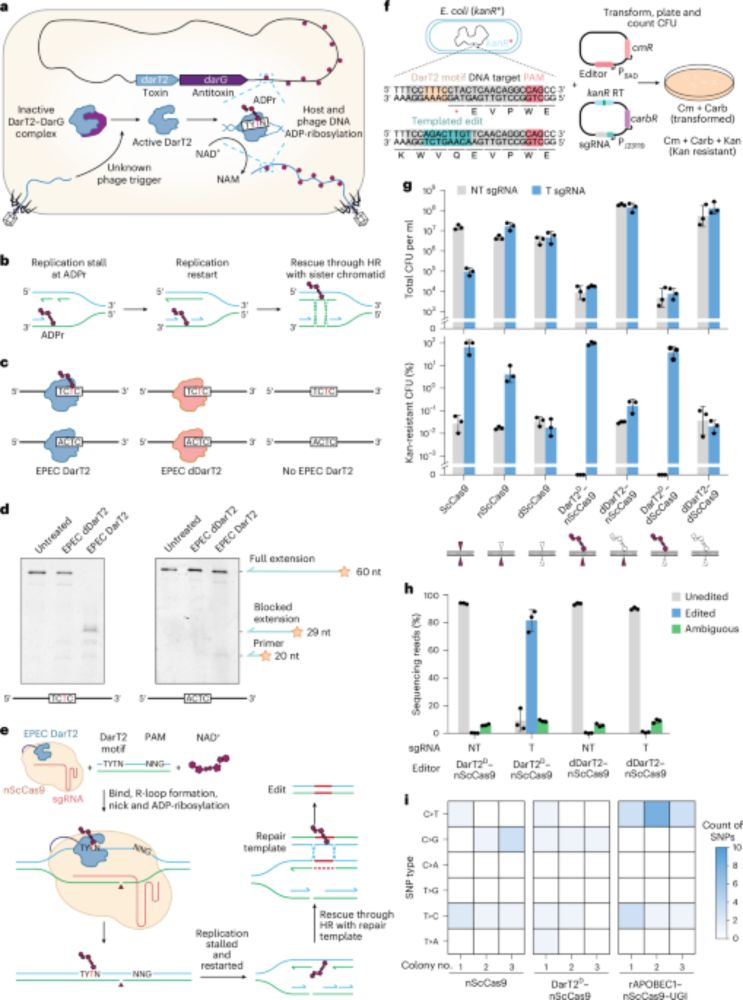
Targeted DNA ADP-ribosylation triggers templated repair in bacteria and base mutagenesis in eukaryotes - @helmholtz-hiri.bsky.social go.nature.com/4n7zDpG
04.09.2025 15:20 — 👍 7 🔁 4 💬 1 📌 0Big shout-out to first authors Darshana Gupta, (myself :) ) & Harris V. Bassett, senior author Chase L. Beisel, and project initiator Scott P. Collins 🙌
04.09.2025 18:51 — 👍 0 🔁 0 💬 0 📌 0This opens up brand-new possibilities for precision editing and studying DNA repair!
04.09.2025 18:51 — 👍 1 🔁 0 💬 1 📌 0We developed append editing, a genome-editing strategy that installs ADP-ribose groups onto DNA to unlock new repair outcomes:
🔹 In bacteria → precise, scar-free edits via homologous recombination
🔹 In eukaryotes → unique T→A or T→C base substitutions not possible with current editors

🚀 New publication alert! 🚀
We just published in @natbiotech.nature.com:
👉 Targeted DNA ADP-ribosylation triggers templated repair in bacteria and base mutagenesis in eukaryotes
doi.org/10.1038/s415...

Learn more ➡️ www.gmc.vu.lt/en/lsc-embl/...
07.05.2025 15:26 — 👍 0 🔁 0 💬 0 📌 0Deadline: Open until a suitable candidate is found.
📧 Send your CV + Motivation Letter to: constantinos.patinios@gmc.vu.lt
🧬 Our work focuses on next-generation genome editing tools for bacteria, yeasts, and phages — with a special focus on DNA replication stalling and repair!
07.05.2025 15:26 — 👍 1 🔁 0 💬 1 📌 0🌍 Based in beautiful Vilnius, Lithuania
🔬 Access to state-of-the-art equipment and facilities
💼 Competitive salary, personalized mentorship, and cutting-edge research!
If you’re passionate about genome editing or synthetic biology…
If you want tailored mentorship to drive independent research and launch your career…
This is the place for you! ✨
🚀 Join the Patinios Lab at the VU LSC - EMBL Partnership Institute for Genome Editing Technologies!
We are looking for a Postdoctoral Researcher to join our growing team! 🔎

Good news on this 35th anniversary of the #Restoration of #Independence in #Lithuania!
A new ERC project from a Lithuanian researcher can now go ahead, as they were able to secure alternative EU funding for her ERC Consolidator Grant.
How did this happen? Find out in the thread 🧵👇
Excited to see our SIBR work out in Trends in Biotechnology :)
doi.org/10.1016/j.ti...
This is the first study I have coordinated from start to finish, and it brings me immense satisfaction to see it published.
For a brilliant description of the tech check this: www.linkedin.com/feed/update/...
Huge thanks to Simona Della Valle, Enrico Orsi and Sjoerd Creutzburg for driving this work.
Happy editing :)

If you want to use SIBR2.0 for your gene/protein of interest, you can visit the SIBR Site Finder site: colab.research.google.com/drive/162gIZ...
25.11.2024 13:47 — 👍 0 🔁 0 💬 1 📌 0SIBR2.0 can virtually control the expression of any gene/protein of interest, in a wide range of (if not all) mesophilic prokaryotes
25.11.2024 13:47 — 👍 0 🔁 0 💬 1 📌 0By using SIBR2.0-Cas12a we achieved ~70% editing efficiency, in just 48h after electroporation
25.11.2024 13:47 — 👍 0 🔁 0 💬 1 📌 0We show that SIBR2.0 is great for inhibiting the translation of the T7 RNAP in E. coli and then we used it to tightly and inducibly express the Cas12a protein in C. necator
25.11.2024 13:47 — 👍 0 🔁 0 💬 1 📌 0his means that even with the alternative translation site present in the SIBR sequence, a non-functional and split protein will be produced
25.11.2024 13:47 — 👍 0 🔁 0 💬 1 📌 0This is where magic happened!! We developed SIBR2.0 :)
SIBR2.0 includes the introduction of SIBR within the coding sequence of the gene of interest, splitting it into 2 distinct exons
The alternative site for translation appeared to be gene- and organism- specific
25.11.2024 13:47 — 👍 0 🔁 0 💬 1 📌 0