Fantastic Gioele!
16.10.2025 07:47 — 👍 1 🔁 2 💬 0 📌 0Noah Holzleitner
@noahholzleitner.bsky.social
PhD Student at Grünewald Lab (TUM) 🧬Protein Design for CRISPR proteins🧑🏽💻👨🏽🎨
@noahholzleitner.bsky.social
PhD Student at Grünewald Lab (TUM) 🧬Protein Design for CRISPR proteins🧑🏽💻👨🏽🎨
Fantastic Gioele!
16.10.2025 07:47 — 👍 1 🔁 2 💬 0 📌 0Super interesting! Love the “Turning an Art into Science” aspect 🧬
15.10.2025 18:52 — 👍 4 🔁 0 💬 0 📌 0Anyone from the Bio community already got their hands on a NVidia DGX Spark for structure prediction and Protein design workloads ?
15.10.2025 05:43 — 👍 3 🔁 2 💬 3 📌 0Mapping the diverse topologies of protein-protein interaction fitness landscapes https://www.biorxiv.org/content/10.1101/2025.10.14.682342v1
15.10.2025 03:03 — 👍 1 🔁 3 💬 0 📌 0
Two group leader positions available in the broader areas of RNA science, RNA technologies, and RNA medicine. Attractive packages and a great environment. Come and join us at Helmholtz RNA Würzburg, Bavaria.
08.10.2025 21:45 — 👍 67 🔁 78 💬 1 📌 1In any case, I am happy that we were lucky enough to receive funding and am excited to be working on the project. To everyone else who have also been rejected multiple times, I wish you the best of luck—if you are lucky enough to be in a position where you can try again. 13/n; n=13
08.10.2025 08:05 — 👍 6 🔁 1 💬 0 📌 0
Directed evolution of a compact TranC11a system for efficient genome editing

Figure 1

Figure 2

Figure 3
Directed evolution of a compact TranC11a system for efficient genome editing [new]
Evolved compact TranC11a matched SpCas9 edit, trait mod in plant/human.

The team is already working to expand the platform’s abilities, including testing in clinically relevant immune and stem cells, and engineering future versions of the system that can rearrange sequences beyond one megabase.
Learn more in the full paper: www.science.org/doi/10.1126/...
Looks very nice!
24.09.2025 16:15 — 👍 2 🔁 0 💬 0 📌 0
Reduction of prime editing errors by engineered Cas9 variants with relaxed nick positioning. #NBThighlight www.nature.com/articles/s41...
22.09.2025 11:54 — 👍 10 🔁 3 💬 1 📌 0very cool work and a milestone in synthetic biology. how impressive are the new phage genomes?
with generative bioML, i'm always looking at how similar the generated sequences are to known sequences. let's take a look

Optimization of a bespoke base editor to treat a severe pediatric vascular disease! 🫀🧬
Our manuscript describes:
1️⃣ Engineering a target-specific BE🧬
2⃣ A *must avoid* bystander edit that occurs with WT SpCas9 BEs! 🙅♂️
3⃣ Extension of lifespan after in vivo editing! 🐁✅
www.nature.com/articles/s41...

Looking for a new approach to studying or eliminating phages? Check out our study introducing anti-phage ASOs (antisense oligos) out in @Nature today. nature.com/articles/s4158…
10.09.2025 15:40 — 👍 133 🔁 65 💬 4 📌 2
Recently tested some de-novo minibinders against two targets (thanks Adaptyv!) designed using our open-source design library, `mosaic`; our best method got hit rates of 7/10 and 8/10 and affinities as low as single-digit nanomolar. Wrote up some thoughts here: blog.escalante.bio/minibinder-d...
02.09.2025 14:39 — 👍 38 🔁 15 💬 2 📌 1Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...

RFdiffusion2 is now live!
github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!

If you use Boltz1/2, BioEmu, Chai1, or other MSA-dependent models, you’re likely using our ColabFold server. Please be considerate! Avoid large submissions across many IPs instead generate the MSA locally. Our server is an old-timer from 2014 and can’t handle that load.
15.08.2025 17:48 — 👍 62 🔁 10 💬 3 📌 0
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Exploring the deletion landscape of S. aureus Cas9 with SABER

Figure 1

Figure 2

Figure 3
Exploring the deletion landscape of S. aureus Cas9 with SABER [new]
SABER maps SaCas9 deletion tolerance for DNA binding, informing design of minimized CRISPRi repressors, revealing insights into SaCas9 structure.

This preprint from Helen Sakharova is one of the coolest things to come out of my lab: “Protein language models reveal evolutionary constraints on synonymous codon choice.” Codon choice is a big puzzle in how information is encoded in genomes, and we have a new angle. www.biorxiv.org/content/10.1...
07.08.2025 08:29 — 👍 214 🔁 83 💬 6 📌 4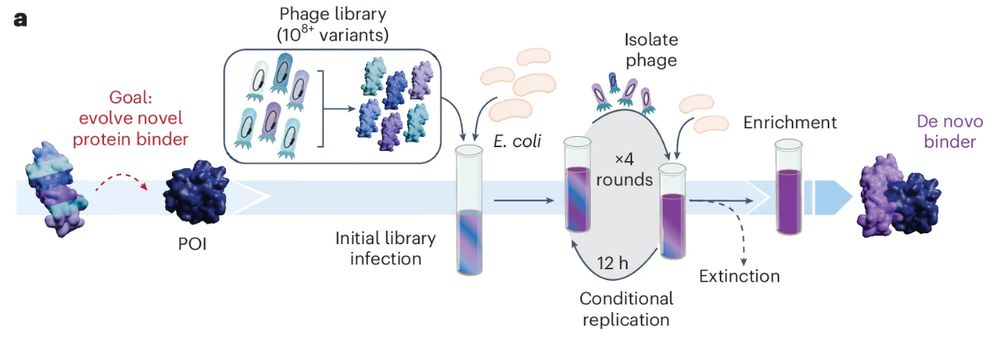
PANCS-Binders (phage-assisted noncontinuous selection of protein binders) screens multiple high-diversity protein libraries against a panel of dozens of targets for high-throughput binder discovery. @chembiobryan.bsky.social @mstyles-chembiol.bsky.social
www.nature.com/articles/s41...
This looks pretty cool to advance protein testing!
06.08.2025 15:38 — 👍 3 🔁 0 💬 0 📌 0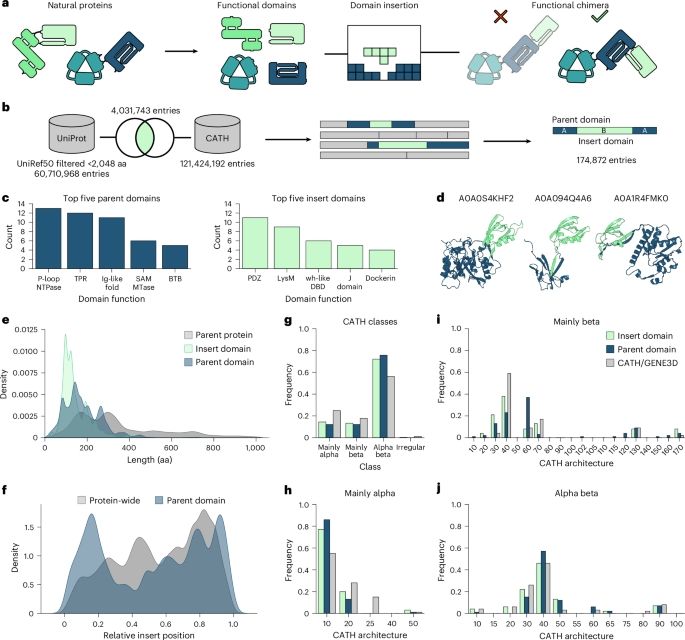
Our work on rationally engineering allosteric protein switches is now out in Nature Methods: www.nature.com/articles/s41... Thanks a lot to @grunewald.bsky.social and @noahholzleitner.bsky.social for the comprehensive news and views: www.nature.com/articles/s41...
04.08.2025 11:41 — 👍 25 🔁 8 💬 0 📌 0
Publication alert: Our paper on domain insertion predictions in proteins is now out in @natmethods.nature.com in its final form: rdcu.be/ey7w3
Also check out the nice perspective by @noahholzleitner.bsky.social and @grunewald.bsky.social : www.nature.com/articles/s41...
Now that OpenCRISPR is in nature and rekindled the 'what's-a-novel-sequence' debate, I'm happy to share an app to check this, which I built for fun some time ago.
fuerstlab.shinyapps.io/SeqNovelty/
quick 🧵


RNAmed students & PIs just returned from the annual retreat on San Servolo, Venice - 3 days of science, collaboration & inspiring talks. Huge thanks to the SAB, including Frank Slack and Michelle Hastings and our speakers Ling-Ling Chen, Li Yang & Gaia Martinelli!
29.07.2025 09:25 — 👍 2 🔁 1 💬 0 📌 0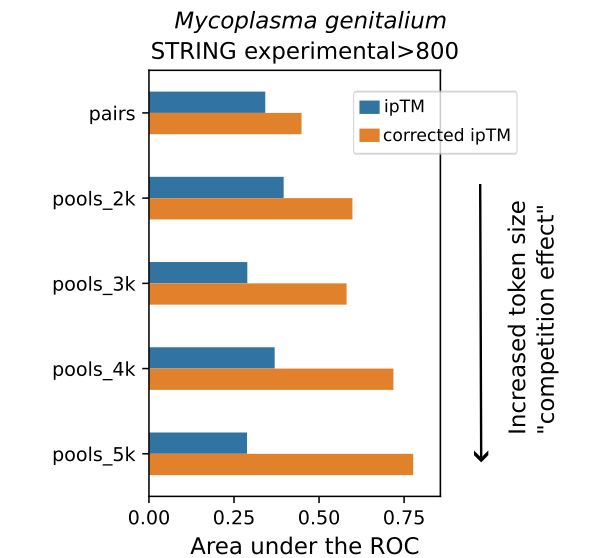
Unexpectedly, @jurgjn.bsky.social found that running Alphafold3 predictions for protein interactions can yield ipTM scores that are more predictive of true interactions when run in pools of proteins instead of pairwise predictions. Presumably, this reflects some sort of "competition effect".
22.07.2025 14:13 — 👍 87 🔁 30 💬 3 📌 3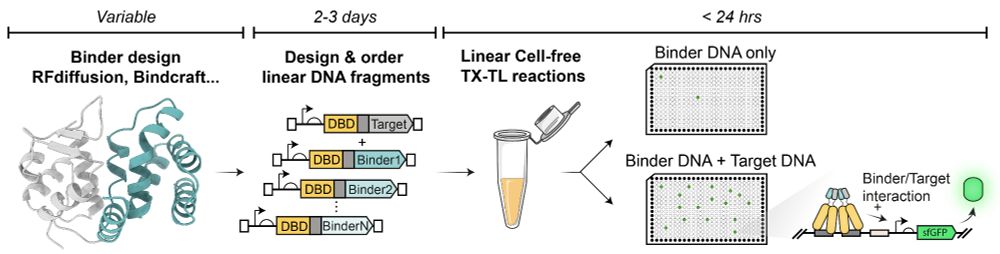
You designed binders for your favourite protein and wish there was a way to experimentally screen them within 24h w/ only a set of pipettes and a plate reader?
Check out our Cell-Free 2-Hybrid approach (CF2H)
Full post: tinyurl.com/48cz5nb6
Preprint: www.biorxiv.org/cgi/content/...
Structural motif search across the protein-universe with Folddisco https://www.biorxiv.org/content/10.1101/2025.07.06.663357v1
07.07.2025 03:48 — 👍 25 🔁 13 💬 0 📌 0
Figure 1

Figure 2

Figure 3

Figure 4
Fold-Conditioned De Novo Binder Design via AlphaFold2-Multimer Hallucination. [new]
FoldCraft: uses AlphaFold2-Multimer with contact map loss to hallucinate novel binders with user-defined folds, improving nanobody design.