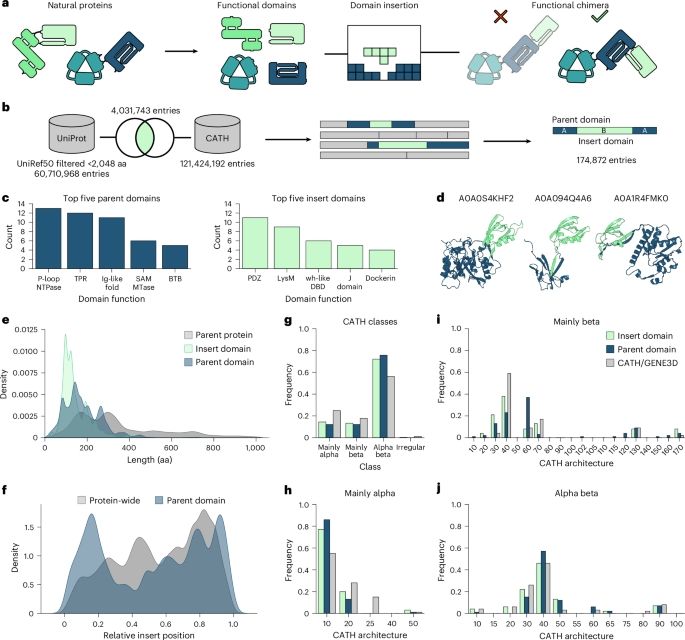
Phage-Assisted Evolution of Allosteric Protein Switches
Allostery, the transmission of locally induced conformational changes to distant functional sites, is a key mechanism for protein regulation. Artificial allosteric effectors enable remote manipulation...
Check out the new pre-print from our lab on phage-assisted evolution of light-switchable, allosteric proteins. Congrats to first author @neuroscinikolai.bsky.social, co-corresponding author @jmathony.bsky.social and everyone from the @niopeklab.bsky.social involved!
www.biorxiv.org/content/10.1...
13.06.2025 10:13 — 👍 21 🔁 7 💬 0 📌 1
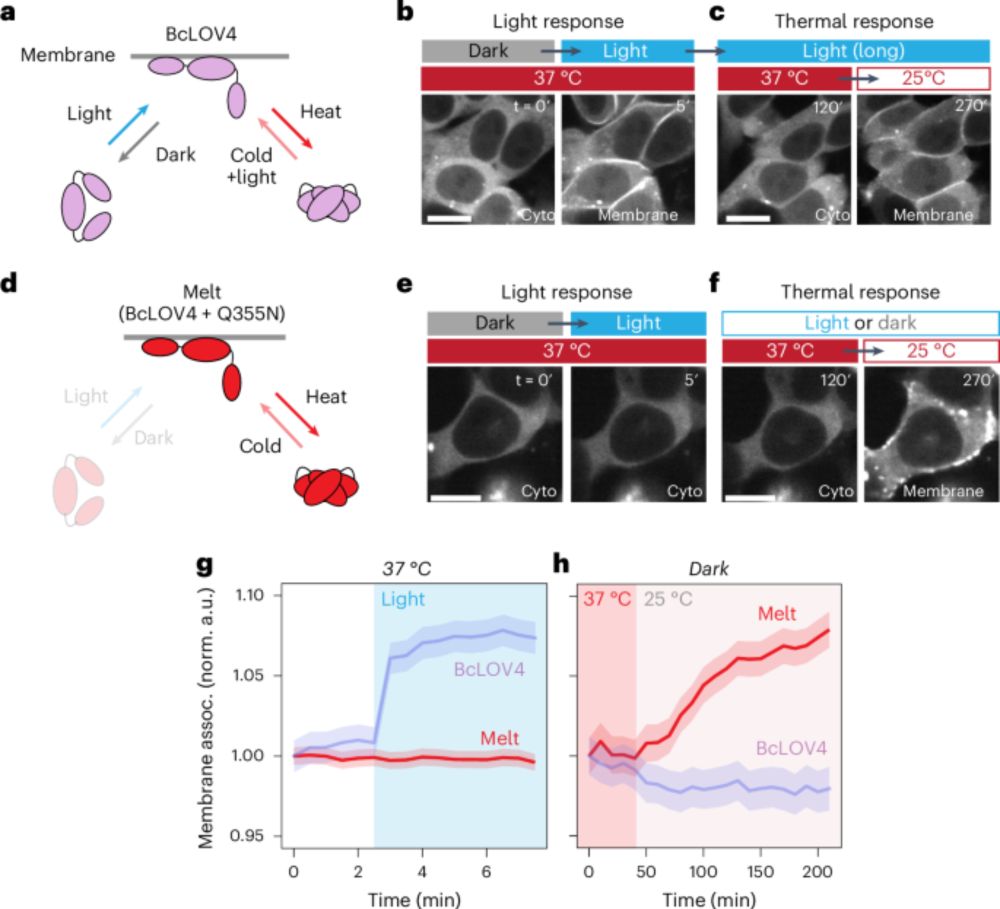
New paper alert! We introduce the modular allosteric thermo-control of protein activity. Employing the AsLOV2 domain and mutants thereof as thermoreceptors, we engineered diverse hybrid proteins, whose activity can be controlled by small temperature changes (37-40/41 °C).
doi.org/10.1101/2025...
03.05.2025 11:03 — 👍 13 🔁 5 💬 3 📌 0

We have an exciting new PhD opportunity!
If you are fsacinated by proteins and their dynamics and want to engineer them to develop new molecular tools: apply now!
We are a young and dynamic team combining state-of-the-art laboratory and bioinformatics approaches.
Please share.
01.04.2025 11:52 — 👍 13 🔁 11 💬 0 📌 1

I'd like to share a little bit of happy lab news in these chaotic times: a new preprint, driven by the brilliant Qinhao Cao!
www.biorxiv.org/content/10.1...
We address a big challenge in synbio: If you give me a protein "X", how can I give you a version of X whose activity is controlled by a kinase?
21.04.2025 16:07 — 👍 78 🔁 31 💬 2 📌 4
I really enjoyed it. That was a fantastic meeting. See you soon in HD @hellmichgroup.bsky.social
01.02.2025 21:47 — 👍 1 🔁 0 💬 0 📌 0
Great to see that thermogenetics starts to take off. Very exciting future ahead for the field, indeed. Congrats @bugajlab.bsky.social for laying important groundwork for others to build on.
24.01.2025 08:44 — 👍 4 🔁 0 💬 1 📌 0
Our paper on computational design of chemically induced protein interactions is out in @natureportfolio.bsky.social. Big thanks to all co-authors, especially Anthony Marchand, Stephen Buckley and Bruno Correia!
t.co/vtYlhi8aQm
15.01.2025 16:37 — 👍 65 🔁 25 💬 1 📌 0
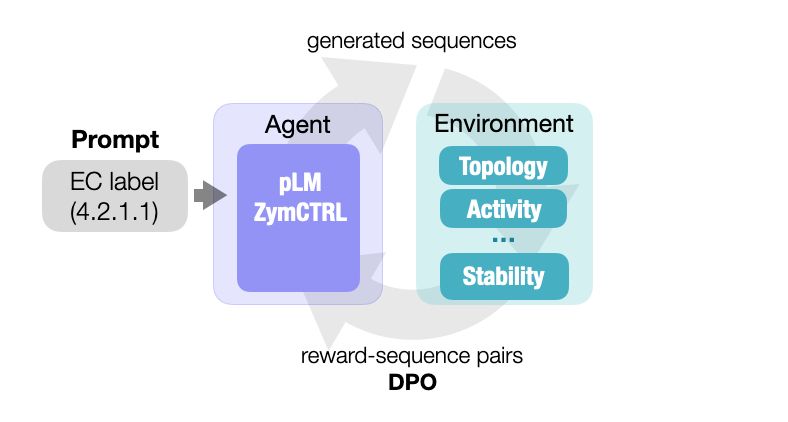
Protein language models excel at generating functional yet remarkably diverse artificial sequences.
They however fail to naturally sample rare datapoints, like very high activities.
In our new preprint, we show that RL can solve this without the need for additional data:
arxiv.org/abs/2412.12979
18.12.2024 21:06 — 👍 58 🔁 15 💬 1 📌 1
We trained a model to co-generate protein sequence and structure by working in the ESMFold latent space, which encodes both. PLAID only requires sequences for training but generates all-atom structures!
Really proud of @amyxlu.bsky.social 's effort leading this project end-to-end!
09.12.2024 14:58 — 👍 57 🔁 11 💬 2 📌 0
Very excited about the first preprint from our lab at TUM! This is a fun gene & cell therapy collaboration led by
@schmidts.bsky.social's lab. We use generative #AI
to design new #CARTcell therapies. Check out Andrea's thread & thanks to the whole team! www.biorxiv.org/content/10.1...
26.11.2024 14:13 — 👍 36 🔁 8 💬 7 📌 0
Congratulations to first author @bene837.bsky.social, co-senior author @jmathony.bsky.social and all co-authors for making this exciting project happen! Comments are highly appreciated
05.12.2024 12:33 — 👍 4 🔁 0 💬 0 📌 0
05.12.2024 12:31 — 👍 4 🔁 1 💬 1 📌 0
Prof at Heidelberg Uni, #Genomics|Program German Cancer Research Center (DKFZ) @Boutroslab @UniHeidelberg @DKFZ @sfb1324 @MKolleg https://michaelboutros.org/about
Posts = personal views
Assistant Professor at @UCSF_BTS | @HHMINEWS HGF | | Loving membrane proteins every day https://www.wcoyotelab.com
Prof TT Uni Bonn & University Hospital Bonn | RNA & ribosome & biochemistry & innate immunology | Postdoc Barna Lab Stanford | PhD Stoecklin Lab DKFZ ZMBH Uni Heidelberg | triplet | https://www.leppeklab.org
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Emmy Noether Research Group Leader Immuno-Materials @ Leibniz INM and @ MaxPlanck Bristol | Combining SynBio and immunology with synthetic cells, biophysics, organoids
The Höfer lab studies the epitranscriptomic mechanisms of gene regulation based on NAD-capped RNAs in bacteria and bacteriophages. We are located at the Max Planck Institute for terrestrial Microbiology, Marburg, Germany and area Member of SYNMIKRO
Physicist, Group Leader @German Cancer Research Center, RNA-Protein Complexes, Cell Division #Riboregulation #RNA #RNAsky #Mitosis #Interdisciplinary #Teaching #Mentoring
www.dkfz.de/en/RNA-Protein-Komplexe-Zellproliferation/index.php
RBP2GO.dkfz.de
RNA biochemist; like RNA structures and RBPs; Marburg University, Germany
Staff Research Engineer in BioAI at InstaDeep (part of @biontech.bsky.social)- machine learning for personalized cancer vaccines, de novo peptide sequencing and signal peptides.
From Belgium 🇧🇪 currently living in Cape Town 🇿🇦
#bioML #TeamMassSpec
Deputy Director of the Institute for Intelligent Biotechnologies at HelmholtzMunich, former editor @naturebiotech.bsky.social, all views expressed here are my own
Investigating virus dynamics, the innate antiviral response and its involvement in carcinogenesis. Part of the DKFZ research program "Immunology, Infection and Cancer". Posts by Marco Binder.
❗️Old 𝕏-Tweets copied to BSKY, please note "original date" tag❗
Assistant Professor at TUM. CRISPR tech dev, cardiovascular gene & cell therapies. https://www.cos.tum.de/en/cos/research/gruenewald-lab/
Immunotherapy, CAR-T, Protein Design, Synthetic biology, Cell Signaling.
Head of Research Group | Structural Biochemistry | LIT
Leibniz Institute for Immunotherapy
https://lit.eu/our-scientists/dr-leo-scheller/
AI for biology | Stanford and Arc Institute
Generative models for protein design.
Group leader at CRG
Assistant Professor at @EPFL in Computer Science and Life Sciences. PostDoc at @Genentech and @Stanford. PhD at @ETH and @BroadInstitute.
www.aimm.epfl.ch
Engineer/scientist, Nobel Prize in Chemistry 2018
I love evolution, enzymes, protein engineering, AI
Linus Pauling Professor at Caltech
CRISPRologist @ UMass Chan Medical School, RNA Therapeutics Institute. Player for Team Hope, Team Reality, and Team Cat. RNA rules.
Associate Professor @ Mass General Hospital & Harvard Medical School
Genome editing / Protein eng. / Molecular medicine 🇨🇦🧬
Kayden-Lambert MGH Research Scholar '23-28
Structural biologist working on 🖥️ protein design, machine learning🤖, crystallography💎, and cryoEM🔬. Avid weirdness connoisseur 🎩