
Slide thanking the protein databank and all the researchers who contributed structures.
At #bps2026 : I loved the special acknowledgement of the @rcsbpdb.bsky.social and the MANY researchers who generated the experimental macromolecular structures that made it possible to build AI models for structure prediction.
23.02.2026 20:44 — 👍 13 🔁 2 💬 0 📌 0
MDAnalysis is participating in #GSoC 2026!
👉 Read our blog post for details on how you can apply to work with MDAnalysis through GSoC: www.mdanalysis.org/2026/02/19/g...
🗓️ Pre-proposals must be submitted by March 9.
❗For transparency and fairness, use public forums for all #GSoC communications.
23.02.2026 16:31 — 👍 5 🔁 4 💬 0 📌 0

Today's President’s Symposium: Communicating the Value of Biophysics in a Changing World features speakers who will discuss perspectives on the current crisis in US investment in science, and approaches for conveying the value of research to human health and prosperity.https://buff.ly/qcRp51M
22.02.2026 18:30 — 👍 5 🔁 2 💬 0 📌 0

Selfie at BPS 2026
I’m at the BPS2026 meeting @biophysicalsoc.bsky.social in San Francisco. Find me to talk science or if you want some @mdanalysis.bsky.social stickers.
22.02.2026 18:36 — 👍 9 🔁 2 💬 0 📌 0
It was a fantastic meeting. It was really great to see so many young and up-and-coming computational scientists all together, showing what they've been working on and learning what they could be doing. I am really happy how @mdanalysis.bsky.social has been bringing people together.
12.11.2025 04:33 — 👍 4 🔁 0 💬 0 📌 0

Our MDAnalysis User Group Meeting 2025 is in full swing in sunny Arizona ! www.mdanalysis.org/pages/ugm2025/
10.11.2025 16:25 — 👍 10 🔁 3 💬 2 📌 1

The official home of the Python Programming Language
TLDR; The PSF has made the decision to put our community and our shared diversity, equity, and inclusion values ahead of seeking $1.5M in new revenue. Please read and share. pyfound.blogspot.com/2025/10/NSF-...
🧵
27.10.2025 14:47 — 👍 6421 🔁 2756 💬 125 📌 452
Release 2.10.0 of MDAnalysis · MDAnalysis
We're happy to announce the new 2.10.0 release of the MDAnalysis library www.mdanalysis.org/2025/10/26/r...
It contains performance improvements, more parallel analysis tools, and the IMDv3 stream reader for directly processing running simulations.
Thanks to 25 contributors, 16 of which were new!
27.10.2025 15:55 — 👍 4 🔁 3 💬 0 📌 0
Shivan Dragon (I was so excited when I pulled it from an Unlimited Starter ... I still play it when I play dragons)
06.08.2025 23:43 — 👍 0 🔁 0 💬 0 📌 0

MDAnalysis User Group Meeting (logo)
Come to the @mdanalysis.bsky.social User Group Meeting at Arizona State University in November. See www.mdanalysis.org/2025/07/15/u... for our keynote speakers and more updates.
Great opportunity for junior researchers to present their work and to network with the MDAnalysis community.
16.07.2025 01:13 — 👍 9 🔁 3 💬 0 📌 0

Bayesian Nonparametric Analysis of Residence Times for Protein–Lipid Interactions in Molecular Dynamics Simulations
Molecular Dynamics (MD) simulations are a versatile tool to investigate the interactions of proteins within their environments, in particular, of membrane proteins with the surrounding lipids. However, quantitative analysis of lipid–protein binding kinetics has remained challenging due to considerable noise and low frequency of long binding events, even in hundreds of microseconds of simulation data. Here, we apply Bayesian nonparametrics to compute residue-resolved residence time distributions from MD trajectories. Such an analysis characterizes binding processes at different time scales (quantified by their kinetic off-rate) and assigns to each trajectory frame a probability of belonging to a specific process. In this way, we classify trajectory frames in an unsupervised manner and obtain, for example, different binding poses or molecular densities based on the time scale of the process. We demonstrate our approach by characterizing interactions of cholesterol with six different G-protein-coupled receptors (A2AAR, β2AR, CB1R, CB2R, CCK1R, and CCK2R) simulated with coarse-grained MD simulations with the MARTINI model. The nonparametric Bayesian analysis allows us to connect the coarse binding time series data to the underlying molecular picture and thus not only infers accurate binding kinetics with error distributions from MD simulations but also describes molecular events responsible for the broad range of kinetic rates.
Ricky Sexton (in collaboration with Mohamadreza Fazel, Maxwell Schweiger and Steve Pressé at ASU) devised a Bayesian nonparametric analysis to robustly and accurately protein-lipid interactions (residence times) from MD simulations – see doi.org/10.1021/acs.... .
22.04.2025 16:40 — 👍 1 🔁 1 💬 0 📌 0

KLAXON 🚨: Indie SAGE has come out of retirement to release a new short report standing up for diversity, equity and inclusion (DEI) in the sciences!
DEI is not just the right thing to do, it *improves* the quality of the science. Please read and share.
independentsage.org/wp-content/u...
1/2
17.03.2025 11:53 — 👍 1070 🔁 502 💬 19 📌 24
You literally cannot tell this incredibly cool part of American history without centering Native American identity. It is an powerful demonstration of how diversity provided tangible military advantages over an enemy committed to White supremacy.
donmoynihan.substack.com/p/whitewashi...
17.03.2025 19:08 — 👍 3787 🔁 1209 💬 47 📌 38
Release 2.9.0 of MDAnalysis · MDAnalysis
MDAnalysis 2.9.0 (and its blog post) is out: www.mdanalysis.org/2025/03/11/r...!
Includes:
* Additional Gromacs/distopia/parallel analysis support
* New "water"/"precision" keywords
🙏 to the release's 10 contributors (3 new), and to @numfocus.bsky.social/@chanzuckerberg.bsky.social for support.
17.03.2025 07:43 — 👍 31 🔁 12 💬 0 📌 0
@biophysicalsoc.bsky.social you need to be sharing at the conference this week like our lives depend on it. Because they do.
Speak up. Stand up. Get out there and protect out democracy
18.02.2025 03:00 — 👍 29 🔁 10 💬 0 📌 0
There are three ways to ultimate success:
The first way is to be kind.
The second way is to be kind.
The third way is to be kind
-Fred Rogers
22.01.2025 21:41 — 👍 14 🔁 3 💬 0 📌 0
Just shut down for good my twitter account. Live's too short to keep social media accounts that have outlived their time.
23.01.2025 00:38 — 👍 5 🔁 0 💬 1 📌 0
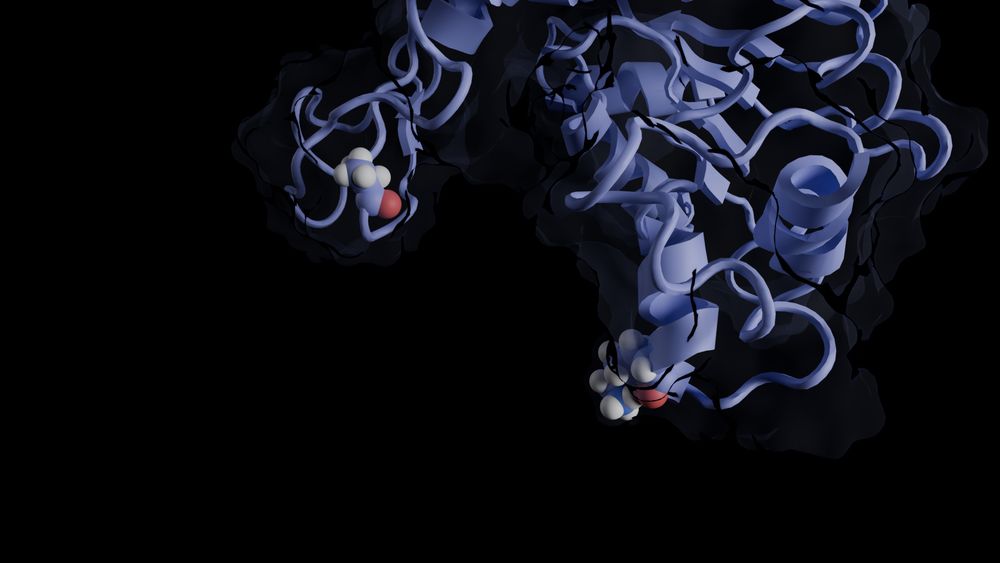
Visualization of part of a protein with MDAnalysis and Blender/MolecularNodes together with Python code to generate the image.
🎉 Small Development Grant by @numfocus.bsky.social for Advanced Molecular Visualizations with #MolecularNodes and @blender3d.bsky.social! @yuxuanzhuang.bsky.social and @bradyajohnston.bsky.social will build an API & integrate with @projectjupyter.bsky.social. See www.mdanalysis.org/2024/12/12/s...
12.12.2024 20:42 — 👍 41 🔁 6 💬 1 📌 0
I had a hard time finding women physicists here. So I made a starter pack for #female and #under-represended #minorities in #physics, in short #FURMIP.
Please spread the word and
let me know if you would like to be added.
go.bsky.app/2JeYsDz
21.11.2024 16:47 — 👍 331 🔁 137 💬 45 📌 9

A reminder that the most productive science is done in collaborations
05.12.2024 17:06 — 👍 67 🔁 12 💬 1 📌 0
GoatCounter – open source web analytics
Goatcounter www.goatcounter.com seems to be doing a decent job for us at mdanalysis.goatcounter.com
27.11.2024 19:50 — 👍 0 🔁 0 💬 0 📌 0
Very pleased that our MDverse project was awarded a prize for Open Science
26.11.2024 18:57 — 👍 37 🔁 9 💬 2 📌 0
How to be a Good Member of a Scientific Software Community [Article v1.0]
| Living Journal of Computational Molecular Science
I'm fuming about another email chain with someone asking me for help with some scientific software I support, so I suppose it's a good time to repost this essay I wrote about how to be a good member of a software community:
livecomsjournal.org/index.php/li...
22.11.2024 16:36 — 👍 31 🔁 9 💬 3 📌 2
Release 2.8.0 of MDAnalysis · MDAnalysis
We released MDAnalysis 2.8.0 🚀 See the blog post www.mdanalysis.org/2024/11/22/r... . Highlights: (1) all code under the GNU Lesser General Public License, (2) new Guesser API, (3) general parallelization for analysis tools, (3) DSSP analysis class, (4) more MDAKits.
27.11.2024 17:33 — 👍 58 🔁 17 💬 0 📌 1
UGM 2024 · MDAnalysis
📢📢 Registration for #mdaUGM2024 is now open! Join us in London from 21-23 Aug to talk about all things MDAnalysis. You also won't want to miss the talks from our keynote speakers, Drs. Antonia Mey and Francesca Stanzione.
Register now to secure your place:
www.mdanalysis.org/pages/ugm202...
17.05.2024 16:30 — 👍 2 🔁 1 💬 0 📌 0

MDAnalysis Workshop June 24-25th 2024, Arizona State University. Stylized saguaro cactus with flowers shaped like the MDAnalysis logo.
Register for our joint MDAnalysis/MolSSI workshop at Arizona State University on June 24th-25th 2024. Attendance is free, travel bursaries are available, register for in-person or online attendance at www.mdanalysis.org/2024/05/14/A... Registration deadline is soon, May 27, 2024.
14.05.2024 21:18 — 👍 2 🔁 1 💬 0 📌 0

MDAnalysis Workshop June 24-25th 2024 at Arizona State University. A Saguaro cactus in the desert with flowers shaped like the MDAnalysis logo.
Save the date: June 24th & 25th: Joint MDAnalysis/MolSSI workshop at Arizona State University. Details to follow.
www.mdanalysis.org/2024/04/24/A...
24.04.2024 18:24 — 👍 2 🔁 1 💬 0 📌 0
Submit your abstract to present at the 2024 @mdanalysis.bsky.social User Group Meeting in London. I so look forward to seeing what everybody is up to!
08.03.2024 20:09 — 👍 3 🔁 2 💬 0 📌 0
Professor of Chemistry@Universidade de Coimbra. Chemistry, cooking, clarinets, symmetry, music, art, computers and programming all make me tick!
https://orcid.org/0000-0003-2393-0203
A group of computational chemists at UCLA. We study catalysts, materials, and complex (bio)molecular systems (and make them better!).
Student-run account.
https://alexandrova.chem.ucla.edu/
Professor of Computational Biochemistry. School of Life Sciences & Department of Chemistry, University of Warwick. Molecular Modeller of Membrane Proteins, Bacterial Biogenesis & Antimicrobial Resistance.
Postdoc in the Cole Group at Newcastle University interested in molecular mechanics force field development and free energy calculations.
Computational chemist, doing CADD @ UCI's Center for Neurotherapeutics. Former chemical engineer turned drug hunter. Buffalonian at heart! Go Bills!!! 🏈
💊🧠💻🧬🧪
#తెలుగోడు
Disclaimer: Views expressed are my own, not those of my employer or affiliates.
Co-Creating Ireland's Public Involvement in Open Research Roadmap
ENGAGED is building a national roadmap to shape public involvement in open research in Ireland. We believe that research can and does play an important role in tackling societal challenges.
We are a computational research group at Newcastle University led by #UKRIFLF Dr Danny Cole specialising in atomistic simulations in medicinal chemistry and biology https://blogs.ncl.ac.uk/danielcole/
Computational Chemist - Protein, peptides, small molecules, crystallization, self-assembly- nanoparticle. Molecular Dynamics, QM/MM, MMPBSA. Dog's mom, interested in music, coffee, food, sports...
Science lead at Open Force Field. Core developer at MDAnalysis. Has never met an opossum.
Mathematics Sorceror (sensory alchemist) at the Arctangent Transpetroglyphics Algra Laboratory (ATAL), I transflarnx mathematics into living rainbows. http://owen.maresh.info https://github.com/graveolensa
Psoeppe-Tlaxtlal, (an undreamt splendour?)
Computational biophysicist, MD research scientist, & German Wednesday enthusiast. 🇮🇪🇬🇧🇪🇺
Aspiring theoretical chemist interested in excited-state dynamics and electronic structure. Passionate teacher. Poet.
Physicist and engineer, into multiple scale models of proteins/RNA/membranes, polymers, scientific computing, more --> http://tinyurl.com/goCompModeling
Computational biophysicist @ Max Planck Institute for Polymer Research, Mainz.
Postdoctoral Researcher @Izmir Biomedicine and Genome Center - Computational Structural Biology Lab.
Scientist, nature-lover, cyclist and climber in my free time
Postdoc in biophysics/biochemistry working on the interaction between the phosphorylated Tau protein and microtubules. Molecular dynamist. Tamer of IDRs and IDPs since 2022
(They/Them)
https://scholar.google.com/citations?user=4S1QUPgAAAAJ&hl=fr
Prof Operational Research , @UCL_CORU, passionate about health care, women in STEMs, defending liberal democracy (!). Member of @independentsage, posts personal. https://www.trumpactiontracker.info/













