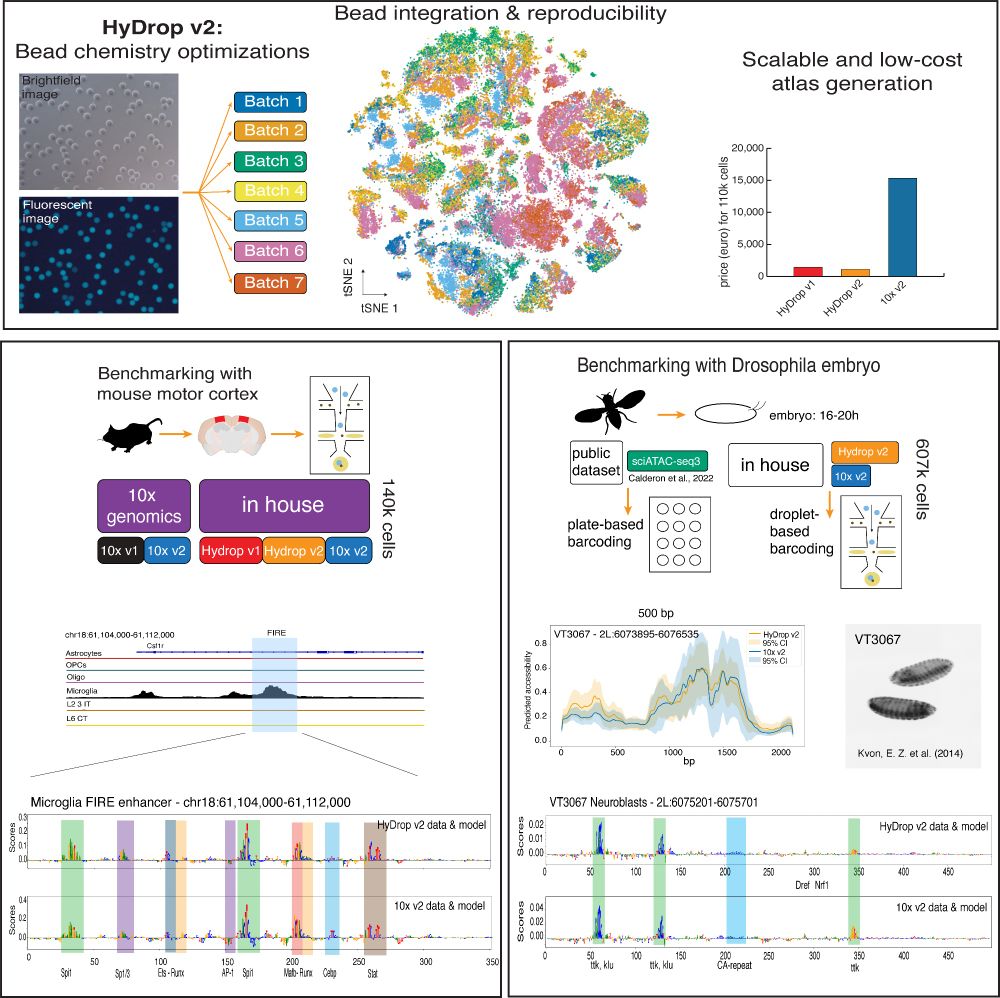
Data collected with the new sequencing platform HyDrop v2 is shown. First, a schematic overview of the bead batches of the microfluidic beads is followed by a tSNE and a barplot showing the costs in comparison to 10x Genomics.
Then, a track of mouse data (cortex) is shown together with nucleotide contribution scores in the FIRE enhancer in microglia. Here, the HyDrop and 10x based models show the same contributions.
On the right, the Drosophila embryo collection is explained; in the paper HyDrop v2 and 10x data are compared to sciATAC data. Then, a nucleotide contribution score is also shown, whereas HyDrop v2 and 10x models show the same contribution, just as in mouse.
Our new preprint is out! We optimized our open-source platform, HyDrop (v2), for scATAC sequencing and generated new atlases for the mouse cortex and Drosophila embryo with 607k cells. Now, we can train sequence-to-function models on data generated with HyDrop v2!
www.biorxiv.org/content/10.1...
04.04.2025 08:52 — 👍 55 🔁 25 💬 2 📌 2
There are 2 mistakes you can make about LLMs:
① Thinking everything LLMs say is correct, they can reason, and with a bit more scale they’ll get us to superintelligence
② Thinking LLMs are good for almost nothing—they are FAR better at all #NLProc tasks than previous methods
12.10.2024 22:38 — 👍 58 🔁 10 💬 1 📌 1
A Practical Guide to Large Scale Docking
Estimated Reading Time: 4 minutes
On this week's, "Deep into the Forest," we cover the practical aspects of running a large scale docking screen such as cleaning up the binding pocket, dealing with conformations, and more deepforest.substack.com/p/a-practica...
23.10.2023 19:18 — 👍 0 🔁 0 💬 0 📌 0
This week on "Deep into the Forest," we explore the use of AlphaFold2 to re-score antibody-antigen complex structure predictions. deepforest.substack.com/p/using-alph...
19.10.2023 03:45 — 👍 5 🔁 0 💬 0 📌 0






