Have you ever wondered how the exact location of a gene affects it's activity?
The main story of my PhD deals with exactly that question, and is now published in Science! ✨
www.science.org/doi/10.1126/...
My amazing co-author and friend @mathiaseder.bsky.social summarized the highlights for you
19.09.2025 14:22 — 👍 36 🔁 13 💬 1 📌 2
Very proud of our paper on "scrambling-by-hopping" LADs, which was just published: www.nature.com/articles/s41.... Congrats to Lise Dauban and the rest of the team – this was a real tour-de-force!
02.09.2025 17:26 — 👍 57 🔁 24 💬 0 📌 0
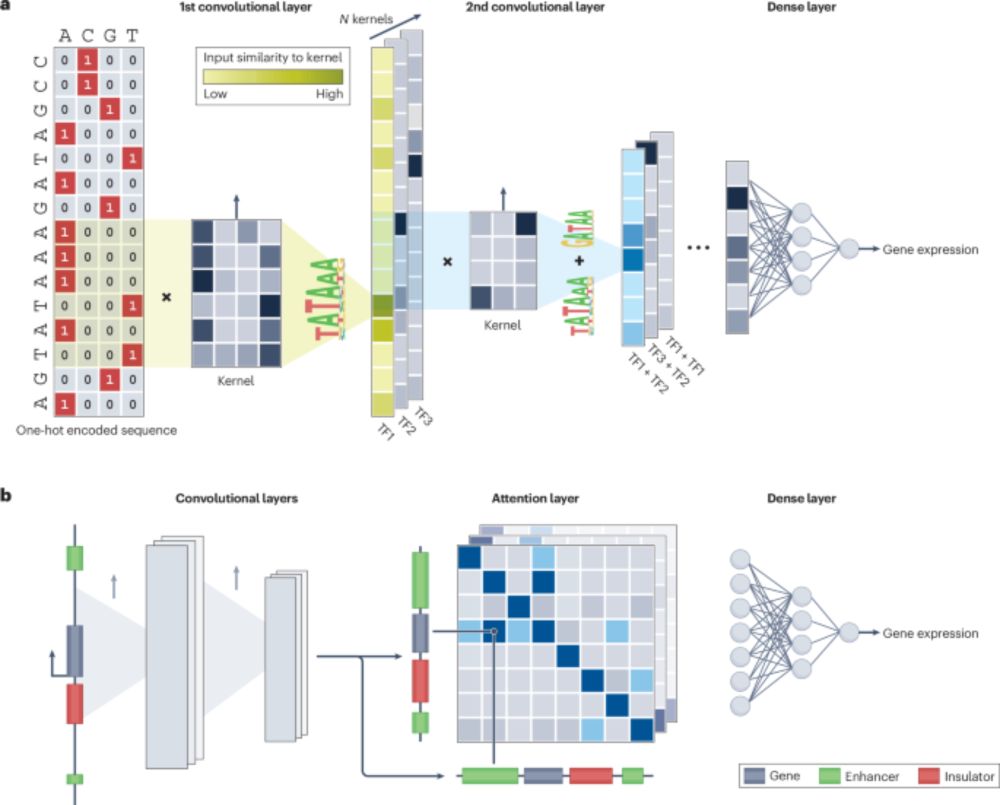
Our review "Predicting gene expression from DNA sequence using deep learning models" is finally out! 🤗
14.05.2025 15:43 — 👍 44 🔁 11 💬 4 📌 2
Welcome to our lab! We are no longer active on Twitter/X. New results/preprints/papers will be posted here.
13.01.2025 22:17 — 👍 10 🔁 3 💬 1 📌 0

A throwback to last month's 'Quantitative biology to molecular mechanisms' conference – time to introduce the poster prize winners! #EMBLOmics
A round of applause for:
🏅 Max Trauernicht
🏅 @ingridpelaez.bsky.social
🏅 Honorine Destain
🏅 Óscar García Blay
Read on 👉🏻 s.embl.org/omx24-01-blog
@embl.org
19.12.2024 14:22 — 👍 9 🔁 4 💬 0 📌 0
Postdoc in the Zwart lab at the Netherlands Cancer Institute (Amsterdam, Netherlands, @nkinl.bsky.social, @zwartlab.bsky.social). Studying gene regulation and 3D-genome organization in cancer.
X/Twitter: @s_gregoricchio
PhD student at @molgen.mpg.de | Schulz Lab - Epigenetics of X-inactivation 🧬🔬
PhD student. Studying gene regulation using AI models | UMCU + NKI, The Netherlands
PhD student in the van Steensel and Medema labs | Netherlands Cancer Institute
GitHub: https://github.com/teunbrand
Postdoc with Bas van Steensel @nkinl.bsky.social
PhD in @noonanlab.bsky.social
Gene regulation, genomics, chromatin, evolution
investigating gene regulation in development and evolution | assistant prof universiteit utrecht | (he)
We are a research lab at the Netherlands Cancer Institute. We develop and apply new genomics tools to study genome biology and gene regulation.
Head of Genome Biology Dept @EMBL,
Scientist, Principal investigator, Professor
Exploring genome regulation during development,
and everything to do with enhancers.
3D genome, chromatin topology,
cell_fate, embryonic Development,
Single Cell genomics
Group Leader @ BIH Berlin and MPI Molecular Genetics. I play around with DNA because I‘d like to know how the genome functions.
Biologist by training, basketball player at heart
Professor for Translational Epigenetics & Genome Architecture, University Medical Center Goettingen, Germany.
www.papantonislab.eu
Lise Meitner Research Group Leader @ Max Planck Institute for Multidisciplinary Sciences
Genome Organisation and Regulation
She/her
Professor of Genetics
Department of Biochemistry
University of Oxford
kloselab.co.uk
Scientist/PI Hubrecht Institute. Exploring gene regulation in early development and cancer. Single-cell spatial genome organization & epigenomics
shendure lab |. krishna.gs.washington.edu
Genomics, AI, sequence-to-function models, mechanisms of the cis-regulatory code. Investigator at the Stowers Institute.
Biochemist and structural biologist with a love for chromatin. Group leader at the Hubrecht Institute.
PI at EPFL / gene regulation / chromatin / epigenetics / genomics / stem cells / transcription factors / single molecule / protein turnover https://www.epfl.ch/labs/suter-lab/ - and 🎹 https://www.youtube.com/@davidsuter594 / https://soundcloud.com/suterd7