ExpansionHunter genotypes are remarkably accurate for both genomes and exomes at the EP400 locus. If you would like to genotype your sample(s), please use the locus definition @ github.com/broadinstitu...
10.01.2025 02:37 — 👍 1 🔁 0 💬 0 📌 0
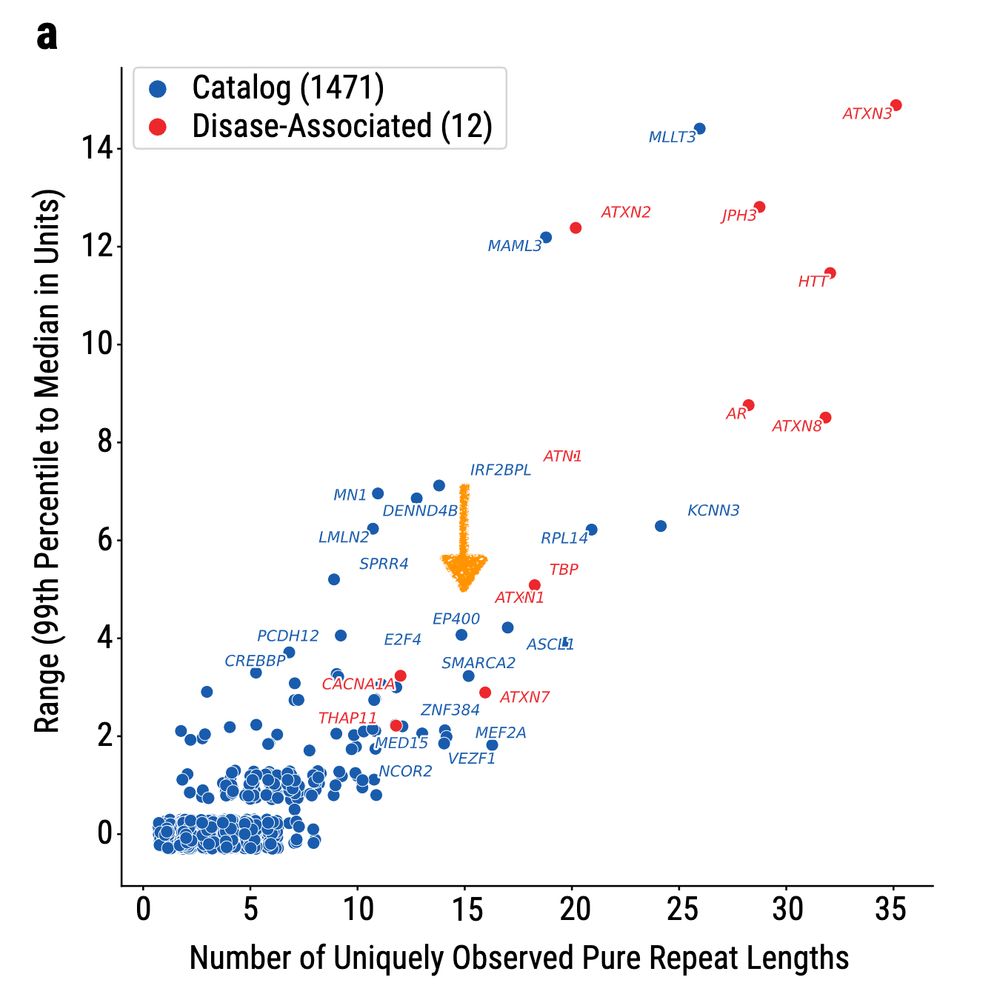
Now, this analysis of 1027 long read AoU genomes has shown that 70% of known disease-associated STR loci are among the top 1% of the most variable STR loci in the genome. Although it wasn't yet a known locus, the EP400 STR also clustered among that top 1%. [5/N]
10.01.2025 02:37 — 👍 0 🔁 0 💬 1 📌 0
Since then, we have genotyped many thousands of other cases and over 1/2 million controls (UKBB, gnomAD, AoU) but haven't found any other samples with more than 50 repeats at this locus, so expansions are extremely rare. [4/N]
10.01.2025 02:37 — 👍 0 🔁 0 💬 1 📌 0
We consider this expansion, found in both affected members of a single family, to be the most likely cause of their spinocerebellar ataxia. [3/N]
10.01.2025 02:37 — 👍 0 🔁 0 💬 1 📌 0
Back in 2022, by genotyping ~5 million STR loci in thousands of short-read rare disease genomes using ExpansionHunter and GangSTR, we discovered an unusually large (≥55 repeats) heterozygous coding CAG expansion in exon #47 of the EP400 gene. [2/N]
10.01.2025 02:37 — 👍 1 🔁 0 💬 1 📌 0
It was great to be able to contribute the EP400 finding to this preprint. Together with @ginaravenscroft.bsky.social and @cfolland.bsky.social we flagged a novel CAG repeat locus in the EP400 gene as a likely (though extremely rare) cause of spinocerebellar ataxia when expanded to ≥55 repeats. [1/N]
10.01.2025 02:37 — 👍 6 🔁 0 💬 1 📌 1
BPS mom, daughter of immigrants, and MBTA commuter working to make Boston a home for everyone.
linktr.ee/mayorwu
Follow the @boston.gov Bluesky Starterpack: https://bsky.app/starter-pack/boston.gov/3lcvbzv3ttb25
Professor of Computer Science @ JHU. https://www.langmead-lab.org/ https://www.youtube.com/BenLangmead
Working towards the safe development of AI for the benefit of all at Université de Montréal, LawZero and Mila.
A.M. Turing Award Recipient and most-cited AI researcher.
https://lawzero.org/en
https://yoshuabengio.org/profile/
MBBS, MD, PhD | GWAS storyteller | Scientist at Regeneron | Human genetics & drug discovery in Neuroscience & Psychiatry
AI @ OpenAI, Tesla, Stanford
I write about medicine at STAT. This is biology's century. Every data point has a face.
Asst. Prof, CU Biomedical Informatics.
#RareDisease #Genomics #STRs #bioinformatics #WomenInSTEM. Parent. Aussie. she/her
Views my own.
https://dashnowlab.org/
Img by familydestinationsguide (CC-BY)
Genomic technologies: WES/WGS, long-read sequencing, optical genome mapping, somatic mutations;
Immuno-genomics: rare diseases; immunodeficiencies; inborn errors of immunity; clonal hematopoiesis
https://shorturl.at/MxQ7O
https://www.immuno-genomics.com
Our mission is to enable the promise of genomics to better human health by creating the world’s most advanced sequencing technologies.
Molecular geneticist | Neurogenetics | repeat expansions | chromosome X | snRNAs | and more
Assistant Investigator @ MGH / Broad / HMS. Focus on human genomics and modeling rare variation. She/her
Professor of Biomolecular Engineering at the University of California, Santa Cruz; Associate Director, UC Santa Cruz Genomics Institute
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Finished a human genome, working on a few more 👨💻
Lab: https://genomeinformatics.github.io
Posts are my own
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
Genomic medicine researcher; chief genomics officer at MGH; clinical lab director at @broadinstitute
MD-PhD candidate at the University of Sydney & Garvan Institute.
Studying tandem repeats in single-cell contexts w/ @dgmacarthur.bsky.social
🧬 Data Scientist at BioAge Labs
🧬 Visiting Scholar at University of Edinburgh
Longevity drug discovery using causal inference, multi-omics, and large language (of life) models
#Longevity #Aging #Lifespan #Healthspan #MachineLearning #AI
