
🥁 NEW
Our study on infant gut microbiome and #straintransmission is now published in @nature.com:
doi.org/10.1038/s415...
1/10
@seandreu.bsky.social
Postdoctoral researcher at @raeslab.org, former Groningen Microbiome Hub. (meta)Genomics, bioinformatics

🥁 NEW
Our study on infant gut microbiome and #straintransmission is now published in @nature.com:
doi.org/10.1038/s415...
1/10

very sad news. Peer Bork was one of the leaders of our field, a wonderful scientist, and he's much too young to be gone. www.embl.org/news/embl-an...
16.01.2026 18:33 — 👍 144 🔁 82 💬 10 📌 7
Grateful to share our paper on gene-specific selective sweeps in human gut microbiomes, now out in Nature! It has been a joy to work with @rwolff.bsky.social, whose insights and hard work made this possible.
www.nature.com/articles/s41...
Had a blast discussing science with Matt David on the Saving Sapiens podcast.
We talk gut microbiome and the findings of our work earlier this year on strain variation linked to host's geography and health, now live on Youtube!
Full episode:
lnkd.in/gUwY36zU
Paper link:
lnkd.in/gNRtqhMF

We selected the laziest mouse at each round to inoculate the next batch of germfree mice: over rounds of selection and passaging, behavior shifted without changes to the mouse genome: rdcu.be/eM3rO
🦠🧫

BIG ANNOUNCEMENT📣: I haven’t been this excited to be part of something new in 15 years… Thrilled to reveal the passion project I’ve been working on for the past year and a half!🙀🥳 (thread 👇)
15.10.2025 12:22 — 👍 492 🔁 185 💬 56 📌 61
#NewResearch
Nasal colonisation by S. aureus is linked with depression in a human cohort and shown in a mouse model to cause decreased serotonin and dopamine in the brain
#MicroSky #Depression
www.nature.com/articles/s41...

Sergio Andreu Sanchez @seandreu.bsky.social (DSM-Firmenich): Understanding the role of the human gut microbiota in aging and health
#HumanMicrobiome
#NIME2025

New Paper!
Machine learning models that attempt to predict microbial load collapse outside of their training context with an R2<0!
In contrast, our Bayesian Partially Identified Models embrace uncertainty in unmeasured microbial load and consistently outpreform.
www.biorxiv.org/content/10.1...
Preprint out for myloasm, our new nanopore / HiFi metagenome assembler!
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N

We present our MetaProViz #Rpackage for #metabolomics analysis & prior knowledge integration to generate mechanistic hypotheses on how metabolic changes affect metabolite classes, pathways & environment interaction
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
The team's first preprint is out!
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
This was seen in totally different human populations (European and Asian), meaning that is unlikely driven by lifestyle common to elders.
Overall we do not know why this is happening, but might highlight a lineage that is better adapted to gut conditions later in life (2/2)
Sure June!
Ruminoccocus gravus is a common member of the gut microbiome. When we looked into the genetics of this bacterium, we saw that older individuals harbored bacteria of similar genetics makeup, while this was not observed in younger people. (1/2)

Do people in the same household share strains when they have the same species?
How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]
🥁 New publication
Our latest study in collab w/ the Groningen Microbiome Hub team is OUT!
We identified 484 microbial-strain-level associations with 241 host phenotypes, encompassing human anthropometric factors, biochemical measurements, diseases, and lifestyle. And more!
For more details 🧵 ⬇️
Big thanks to the researchers and participants behind the studies we used, the Curated curatedMetagenomicData team for making these datasets accessible, and Lifelines for providing cohort and phenotypic data. 🙏 (11/n)
30.04.2025 16:15 — 👍 1 🔁 0 💬 0 📌 0Special thanks to Nicola for hosting me in his lab in @CIBIO @CIBIO_UniTrento—an inspiring few months of great science! Grateful to @embo.org EMBO for funding this experience. (10/n)
30.04.2025 16:15 — 👍 0 🔁 0 💬 1 📌 0@jingyuan-fu.bsky.social bsky.social @daomingwang.bsky.social @vheidrich.bsky.social ch.bsky.social @mireiavallesc.bsky.social allesc.bsky.social @fackelmeister.bsky.social @kmci.bsky.social @umcgroningen.bsky.social (9/n)
30.04.2025 16:15 — 👍 1 🔁 0 💬 1 📌 0This has been a fantastic collaboration between the Groningen Microbiome Hub team and @cibiocm.bsky.social. Huge thanks to all the amazing team members from both groups! (8/n)
30.04.2025 16:15 — 👍 1 🔁 0 💬 1 📌 0While in this work we are unable to assess what is the cause of such associations, we highlight the intriguing links between bacterial strains and human phenotypes.
We provide a list of strain-phenotype associations and large phylogenies, hoping they’ll spark further interest in the community. (7/n)

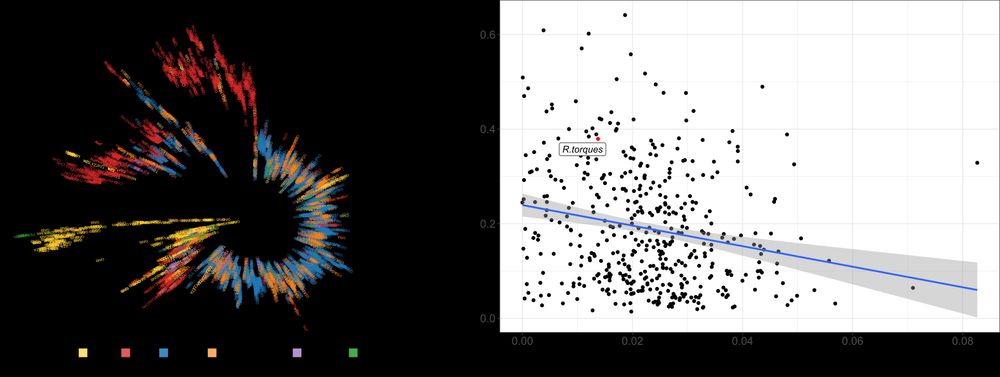
C. aerofaciens phylogeny showed a melanoma-enriched clade across diverse studies.
Interestingly, individuals with prostate cancer also clustered here! (6/n)
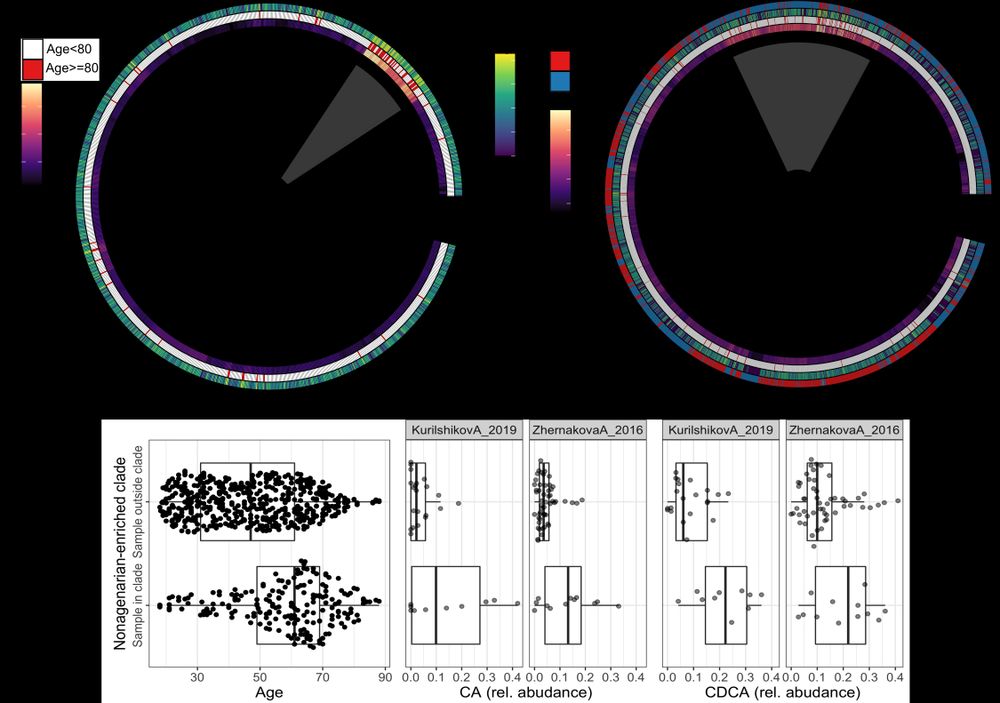
The phylogeny of R. gnavus displayed a clade most commonly seen in older individuals. We observed this clade in both Asian and European studies, and found a distinct bile acid profile in those individuals harboring this R. gnavus clade. (5/n)
30.04.2025 16:15 — 👍 2 🔁 1 💬 2 📌 0Human phenotypes also clustered with species phylogenies.
Association analysis revealed multiple phenotype–species links, including strong, reproducible associations between R. gnavus and age, and C. aerofaciens and melanoma. (4/n)
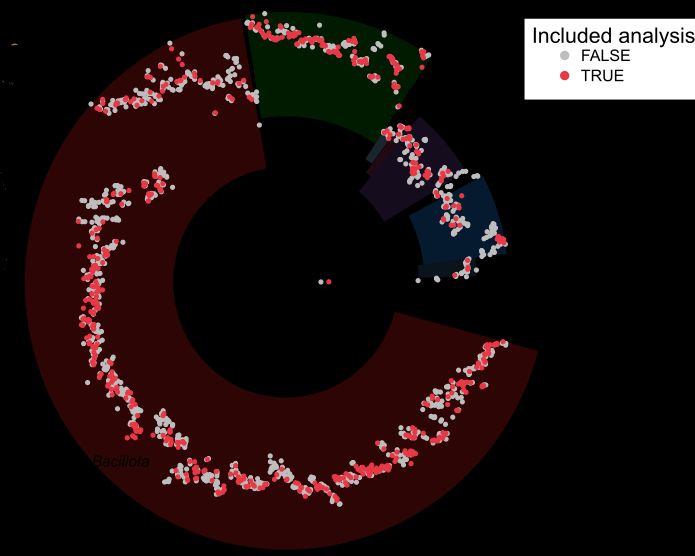
Phylogenies revealed geographic stratification 🌎.
Species with stronger stratification were less often shared across individuals, suggesting limited dispersal drives the observed geographic patterns. (3/n)
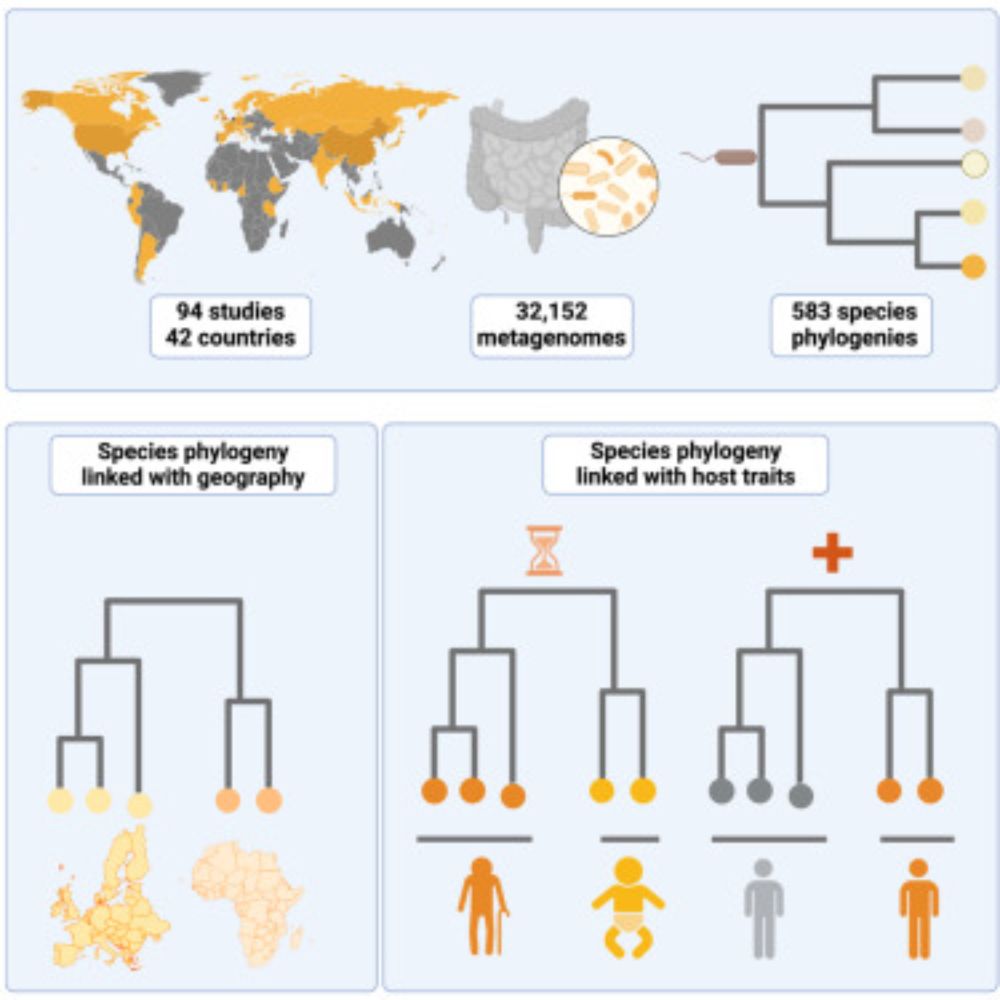
Strain-level genetic variation can shape host–microbe interactions 🦠🧬 .
We analyzed >32,000 public metagenomes, profiled species marker genes at single-nucleotide resolution with StrainPhlAn, and built ~600 species phylogenies. (2/n)
Thrilled to share that our manuscript on strain-level gut microbiome variation across diverse populations and human phenotypes is out today in @cellpress.bsky.social
Curious about how strain diversity relates to human traits? Follow this thread! 🌍 (1/n)
www.sciencedirect.com/science/arti...

After 4,5 years, working besides great colleagues and mentors, last week I received my PhD degree at @unigroningen.bsky.social
I feel really lucky to have had the opportunity to work in such fantastic scientific environment.
Exciting things to come in the next chapter!
What are some classic papers in phylogenetics?
Please repost

Fairy: fast approximate coverage for multi-sample metagenomic binning
www.biorxiv.org/content/10.1...
From
@jimshaw.bsky.social
Finally the answer for those routinely running 40,000 bowtie2 metagenome mappings per project (niche but real)