Range extender mediates long-distance enhancer activity - Nature
The REX element is associated with long-range enhancer–promoter interactions.
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
02.07.2025 16:17 — 👍 186 🔁 90 💬 10 📌 9
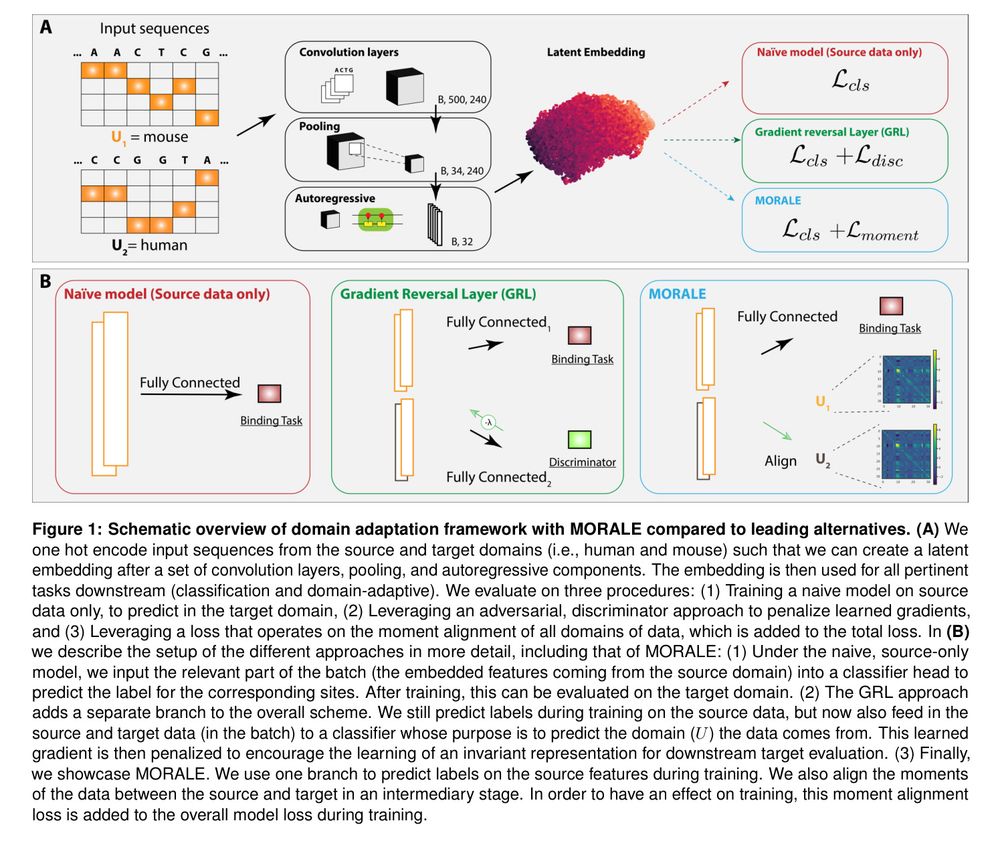
Figure 1
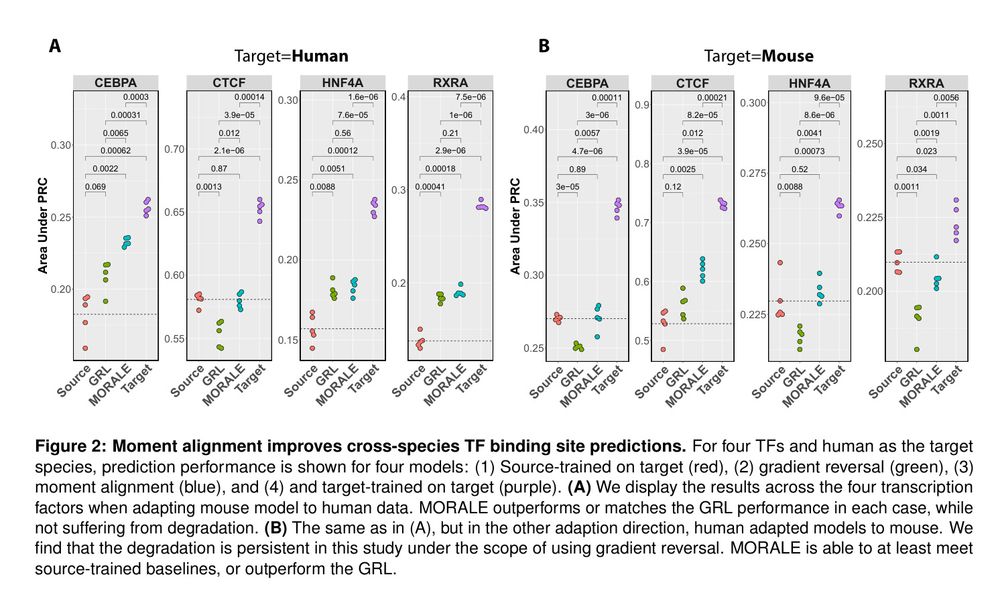
Figure 2
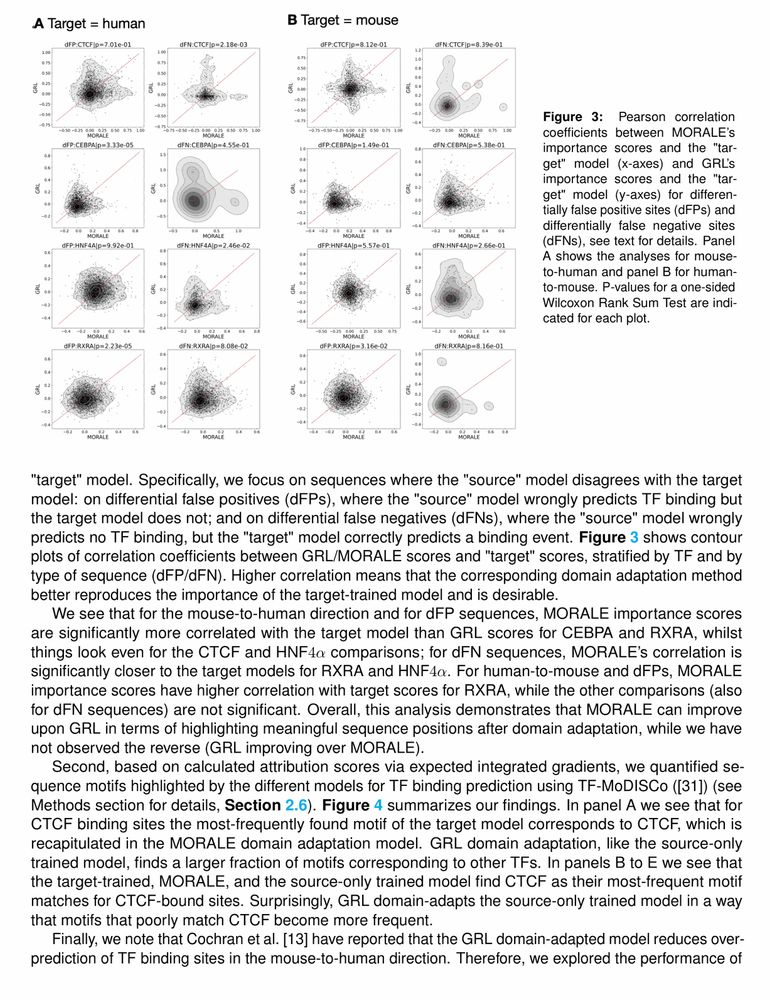
Figure 3
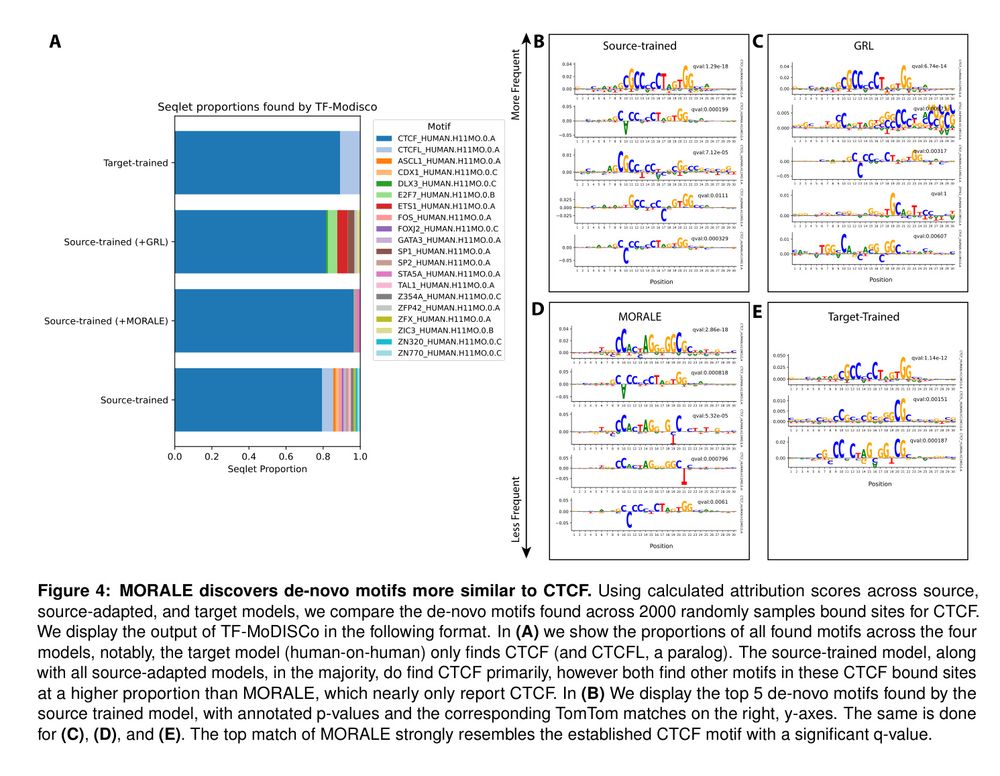
Figure 4
Frustratingly easy domain adaptation for cross-speciestranscription factor binding prediction [new]
Predicts TF binding in target species via aligned sequence data distribution moments for cross-species generalization.
26.05.2025 22:54 — 👍 2 🔁 1 💬 0 📌 0
breaking news, white steam emerges from the Autoclave
08.05.2025 16:23 — 👍 467 🔁 94 💬 6 📌 6

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
Out in Cell @cp-cell.bsky.social: Design principles of cell-state-specific enhancers in hematopoiesis
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵
08.05.2025 16:06 — 👍 140 🔁 58 💬 4 📌 8
If they behave anything like the introverts I know, we won't have to worry about robo spam callers anymore because the LLMs will have to psyche themselves up for hours before they can make the call.
07.05.2025 19:59 — 👍 2 🔁 0 💬 0 📌 0
Many of you enjoy our sequence-based model of single-cell RNA and ATAC data scooby... Don't miss Laura Marten's talk at the upcoming Kipoi seminar about it this Wed!
@lauradmartens.bsky.social @johahi.bsky.social @kipoizoo.bsky.social
Last preprint version:
www.biorxiv.org/content/10.1...
05.05.2025 16:26 — 👍 11 🔁 4 💬 0 📌 0
Hopefully this'll make our models a bit more accessible/usable for all the PyTorch folks out there (including me now 😂).
05.05.2025 20:10 — 👍 0 🔁 0 💬 0 📌 0
(1) small tweaks to bpnetlite's BPNet class to reduce memory footprint & enable training on large, multi-individual datasets and (2) a full reimplementation of CLIPNET in PT using BPNet as its backbone and w/ 2114 bp context length (instead of the original's 1 kb).
05.05.2025 20:09 — 👍 1 🔁 0 💬 1 📌 0
Read the prompt again. I guess some of those papers aren't directly related for gene expression specifically, but successes in epigenomic VEP should be part of the discussion of gene expression VEP IMO.
04.05.2025 23:40 — 👍 1 🔁 0 💬 0 📌 0
It's missing a bunch of important papers too. Fine-tuning work from Ioannidis & Pollard labs, DeepAllele, BigRNA, ChromBPNet, my stuff (obvious COI but it's relevant). Not a cutoff date issue since it's got the pretty recent review from Ioannidis lab.
04.05.2025 23:37 — 👍 1 🔁 0 💬 1 📌 0
New preprint from the @arnausebe.bsky.social lab! 💐
Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.
19.03.2025 10:31 — 👍 57 🔁 27 💬 3 📌 0
Positively transformative *raises pinky*
01.05.2025 22:43 — 👍 0 🔁 0 💬 0 📌 0
I guess that's true, but using language models to learn a cell state autoencoder feels a bit overkill to me.
01.05.2025 20:39 — 👍 2 🔁 0 💬 1 📌 0
It's also not clear to me why we should expect MLM to work for gene expression vectors? Like at least for PLMs and DNALMs I can see the language analogy (and PLMs actually work) and how we might learn something from MLM training. What's being learned from ordered gene expression vectors?
01.05.2025 18:46 — 👍 3 🔁 0 💬 1 📌 0
🎉🎉🎉
29.04.2025 01:15 — 👍 0 🔁 0 💬 0 📌 0

Programmatic design and editing of cis-regulatory elements
The development of modern genome editing tools has enabled researchers to make such edits with high precision but has left unsolved the problem of designing these edits. As a solution, we propose Ledi...
Our preprint on designing and editing cis-regulatory elements using Ledidi is out! Ledidi turns *any* ML model (or set of models) into a designer of edits to DNA sequences that induce desired characteristics.
Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
24.04.2025 12:59 — 👍 115 🔁 37 💬 2 📌 3

High-resolution reconstruction of cell-type specific transcriptional regulatory processes from bulk sequencing samples
Biological systems exhibit remarkable heterogeneity, characterized by intricate interplay among diverse cell types. Resolving the regulatory processes of specific cell types is crucial for delineating developmental mechanisms and disease etiologies. While single-cell sequencing methods such as scRNA-seq and scATAC-seq have revolutionized our understanding of individual cellular functions, adapting bulk genome-wide assays to achieve single-cell resolution of other genomic features remains a significant technical challenge. Here, we introduce Deep-learning-based DEconvolution of Tissue profiles with Accurate Interpretation of Locus-specific Signals (DeepDETAILS), a novel quasi-supervised framework to reconstruct cell-type-specific genomic signals with base-pair precision. DeepDETAILS’ core innovation lies in its ability to perform cross-modality deconvolution using scATAC-seq reference libraries for other bulk datasets, benefiting from the affordability and availability of scATAC-seq data. DeepDETAILS enables high-resolution mapping of genomic signals across diverse cell types, with great versatility for various omics datasets, including nascent transcript sequencing (such as PRO-cap and PRO-seq) and ChIP-seq for chromatin modifications. Our results demonstrate that DeepDETAILS significantly outperformed traditional statistical deconvolution methods. Using DeepDETAILS, we developed a comprehensive compendium of high-resolution nascent transcription and histone modification signals across 39 diverse human tissues and 86 distinct cell types. Furthermore, we applied our compendium to fine-map risk variants associated with Primary Sclerosing Cholangitis (PSC), a progressive cholestatic liver disorder, and revealed a potential etiology of the disease. Our tool and compendium provide invaluable insights into cellular complexity, opening new avenues for studying biological processes in various contexts. ### Competing Interest Statement The authors have declared no competing interest.
www.biorxiv.org/content/10.1...
10.04.2025 17:06 — 👍 1 🔁 0 💬 0 📌 0
Does anyone know if the ATAC-seq bam files on ENCODE have had their tags shifted? and if so, by the more common +4/-5 or by +4/-4?
17.03.2025 01:14 — 👍 0 🔁 0 💬 0 📌 0
My next pull request will make wingdings the default font for sequence logos.
16.03.2025 02:17 — 👍 3 🔁 0 💬 1 📌 0
Kipoi
Join us for our next Kipoi Seminar with with Alexander Sasse
@lxsasse.bsky.social
@zmbh.uni-heidelberg.de
👉Advanced training strategies for genomic sequence-to-function models
📅 Wed March 5, 5:30pm CET
🧬 kipoi.org/seminar/
🦋 @kipoizoo.bsky.social
01.03.2025 19:26 — 👍 14 🔁 6 💬 0 📌 0
ralphi: a deep reinforcement learning framework for haplotype assembly
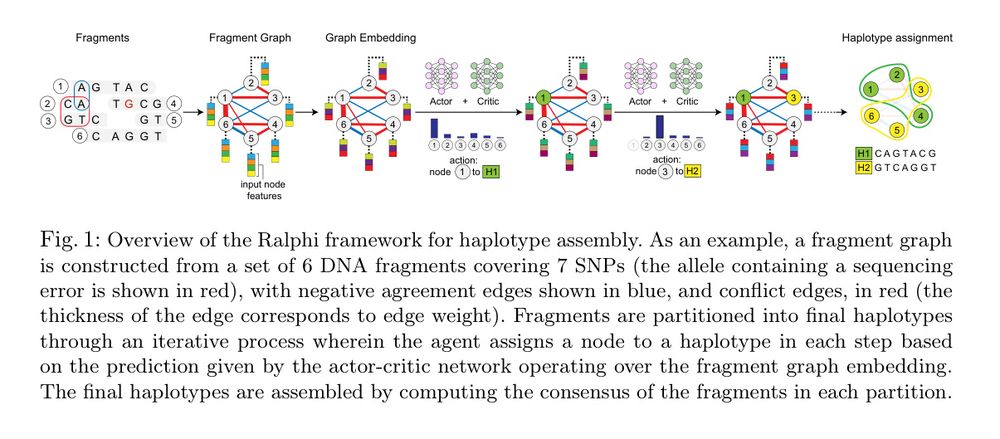
Figure 1
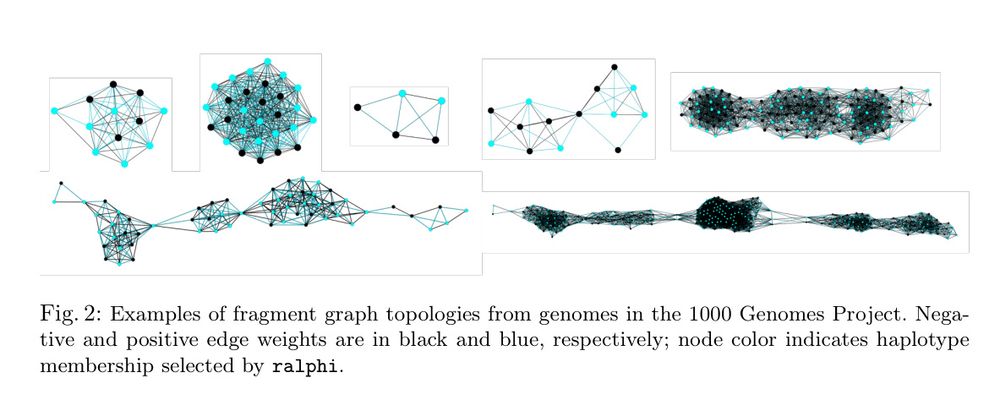
Figure 2
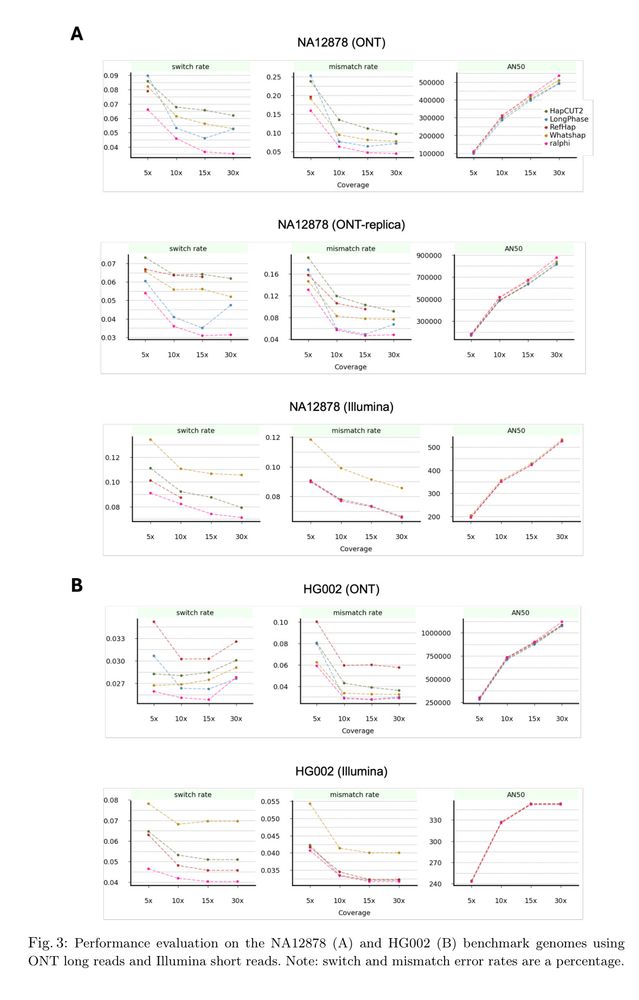
Figure 3
ralphi: a deep reinforcement learning framework for haplotype assembly [new]
Deep reinforcement learning accurately partitions reads into haplotype sets. It uses fragment graphs and the max-cut problem for the reward objective.
22.02.2025 03:50 — 👍 2 🔁 1 💬 0 📌 0

A scalable approach to investigating sequence-to-expression prediction from personal genomes
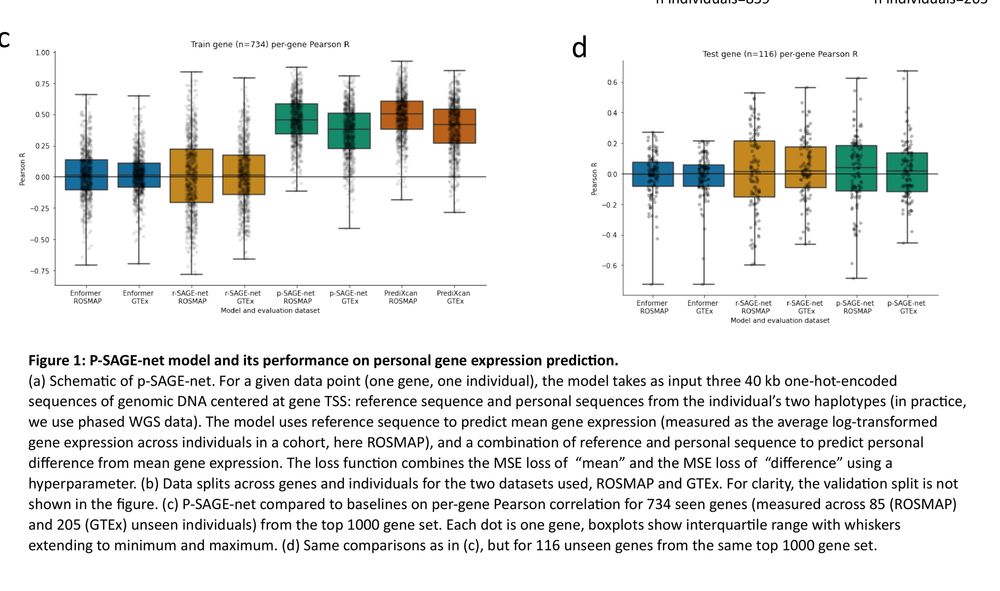
Figure 1
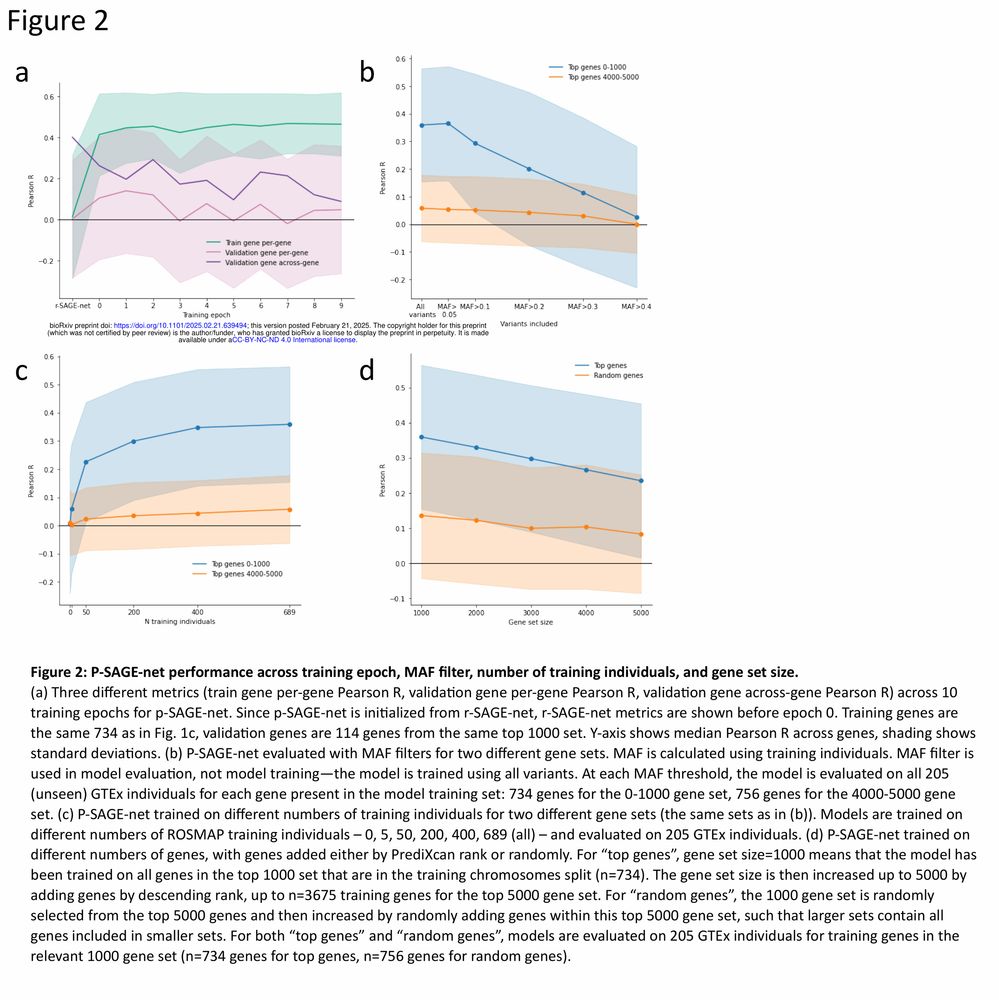
Figure 2
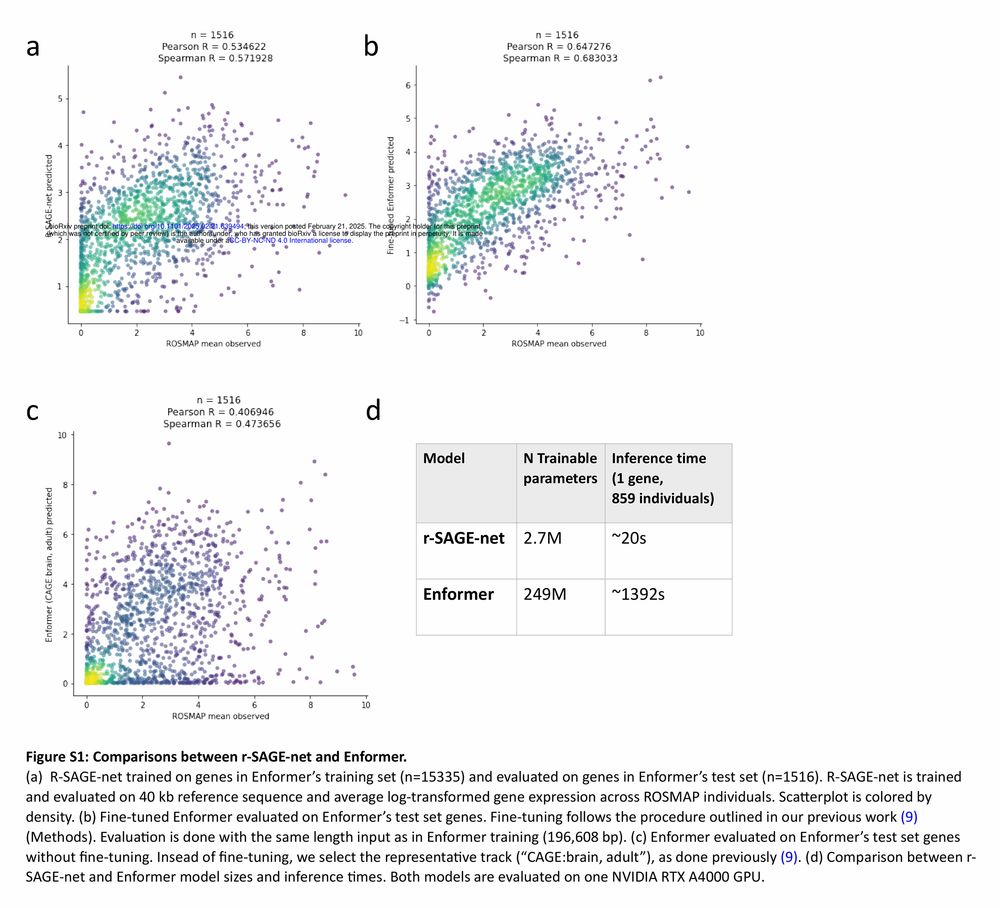
Figure S1
A scalable approach to investigating sequence-to-expression prediction from personal genomes [new]
Models fail to gen. w/ individual var., personal genome training helps some individuals only.
22.02.2025 06:34 — 👍 3 🔁 2 💬 0 📌 0
YouTube video by Bethesda Softworks
Skyrim: Very Special Edition – Official Trailer
The same people who want to play Skyrim on their smart fridge
www.youtube.com/watch?v=FnEW...
19.02.2025 17:46 — 👍 1 🔁 0 💬 1 📌 0
PhD student @gagneurlab.bsky.social (TU Munich and Helmholtz Munich).
Interested in rare variant genetics.
https://shubhankarlondhe.github.io/
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, AI/ML in biotech // http://albertvilella.substack.com
Our long-term research goal is to understand and predict gene regulation based on DNA sequence information and genome-wide experimental data.
Scientist and medical doctor. Biology AI/ML methods, gene regulation, DNA sequence models, single cells. Doing a PhD in computational biology at @molgen.mpg.de.
Computing all things single-cell | Postdoc at Genentech @MarioniLab | Steering council at scverse.org
Engineering cells to understand their decisions
ESPOD Fellow @ebi.embl.org & @sangerinstitute.bsky.social
(Saez-Rodriguez & Parts)
lingering scientist @crick.ac.uk (Briscoe)
Data Scientist Generative AI @BayerCropScience. ML for Plant Biology. PhD @IowaStateUniversity https://www.linkedin.com/in/koushik-nagasubramanian/
Assistant Professor at NYU Langone. Genetics, evolution and biology of complex traits and diseases.
3D genome, chromatin regulation, multi-omic TechDev @ New York Genome Center 🗽, via WEHI 🇦🇺
Assistant Prof at Columbia || Studying data science and its interactions with human behavior || Applications in education, healthcare, and online platforms.
https://hannahql.github.io/
Bioinformatics PhD Candidate @ UMich
CS PhD Candidate at Stanford. Working at the intersection of Machine Learning, Regulatory Genomics, and Complex Disorders
Structural biologist and biochemist. CNRS researcher at CBM Orléans @cbm-upr4301.bsky.social. Interested in protein modifications & interactions. Also husband, dad of 2, friend, ☧. Personal website: msuskiewicz.github.io
PhD student @uwcse; ML&CompGenomics&GeneRegulation; Bio&CS undergrad
@PKU1898; Intern@Yale, Microsoft Research Asia, Genentech
PhD student @ Koo lab, CSHL | Computational Biology | AI Alignment
fly person, PhD student @ Stein Aerts Lab of Computational Biology, VIB.AI
Scientist at University of Edinburgh. Director of MRC Human Genetics Unit but posting in my own capacity.
Human geneticist studying cancer at MSK. Prev. postdoc at Stanford and BS + PhD at Cornell. avid skier, runner, and Yankee fan.
Interested in AI in Genomics - Computational Geneticist @InstaDeep
Ray and Stephanie Lane Professor of Computational Biology at CMU School of Computer Science. https://www.cs.cmu.edu/~jianma/