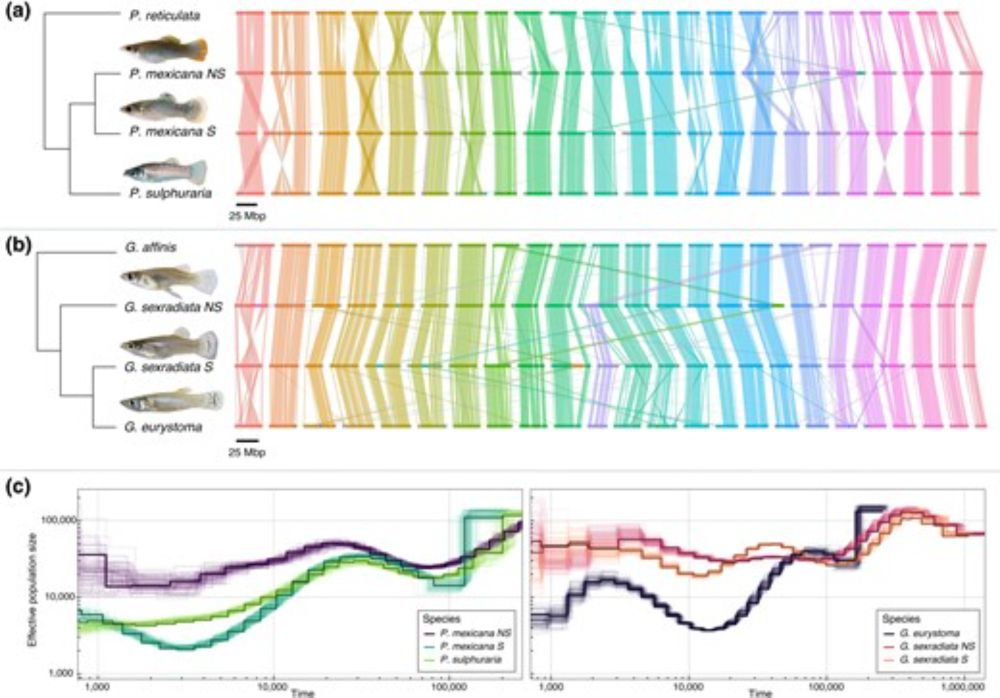
This framework could help us move from serendipitous discovery toward predictive identification of bio-utility across the tree of life. Read the full perspective for the details. [8/8]
05.02.2026 22:02 — 👍 0 🔁 0 💬 0 📌 0
Evolution tells us not just what's useful, but where to look next, highlighting lineages that might harbor untapped potential for biotechnology applications. [7/8]
05.02.2026 22:02 — 👍 0 🔁 0 💬 1 📌 0
But protein data alone isn't enough. Integrating evolutionary relationships between species with this protein information helps us understand where useful functions evolved, distinguish conservation from convergence, and spot breakthrough novelty in constrained protein families. [6/8]
05.02.2026 22:02 — 👍 0 🔁 0 💬 1 📌 0
The key? Identifying indicators of bio-utility from protein variation itself:
• Proteins with extreme properties (like extremophile polymerases)
• Proteins with discrete novel functions (like different proteases)
• Proteins showing conserved or convergent functions (like antifreeze proteins) [5/8]
05.02.2026 22:02 — 👍 0 🔁 0 💬 1 📌 0

Schematic of information that can be leveraged for finding and interpreting bio-utility, including a gene tree, the characteristics of proteins simplified into a 2D protein surface, and the evolutionary relationships between the species in which the same proteins evolved, highlighting the wealth of knowledge and broader perspective we gain about proteins when we include their evolutionary information.
We propose an evolution-integrated framework that combines the best of both worlds: expanding sampling breadth while more deeply interrogating existing data through an evolutionary lens. [4/8]
05.02.2026 22:02 — 👍 1 🔁 0 💬 1 📌 0

Biology needs to become prospective
Since biological data are often non-independent, more data doesn't always mean more insight. We argue that a prospective approach is needed to uncover the deepest principles of life.
In silico bioprospecting tries to fix this by mining existing databases. But those databases are taxonomically biased (see another one of our pubs dropping today: thestacks.org/publications...), and the methods often require knowing what you're looking for upfront. [3/8]
05.02.2026 22:02 — 👍 1 🔁 0 💬 1 📌 0
Bioprospecting has given us incredible discoveries, from life-saving drugs to agricultural innovations. But traditional approaches rely heavily on serendipity and are hard to scale. [2/8]
05.02.2026 22:02 — 👍 1 🔁 0 💬 1 📌 0
Interested in whether protein language models can learn about evolution and leverage this to predict protein structures? Check out the pub!
30.01.2026 23:11 — 👍 1 🔁 0 💬 0 📌 0
I'm very excited to share that today is my last day working at UC Berkeley before I start my new position at @arcadiascience.com next week! It has been a pleasure working as a postdoc in so many amazing labs along side amazing scientists, and I can't wait to start this new chapter!
31.10.2025 16:17 — 👍 7 🔁 0 💬 1 📌 0
I am recruiting PhD students to join my lab at MSU starting Fall 2026! Please spread the word and reach out if you are interested and/or want to learn more!
08.07.2025 13:24 — 👍 13 🔁 15 💬 1 📌 2
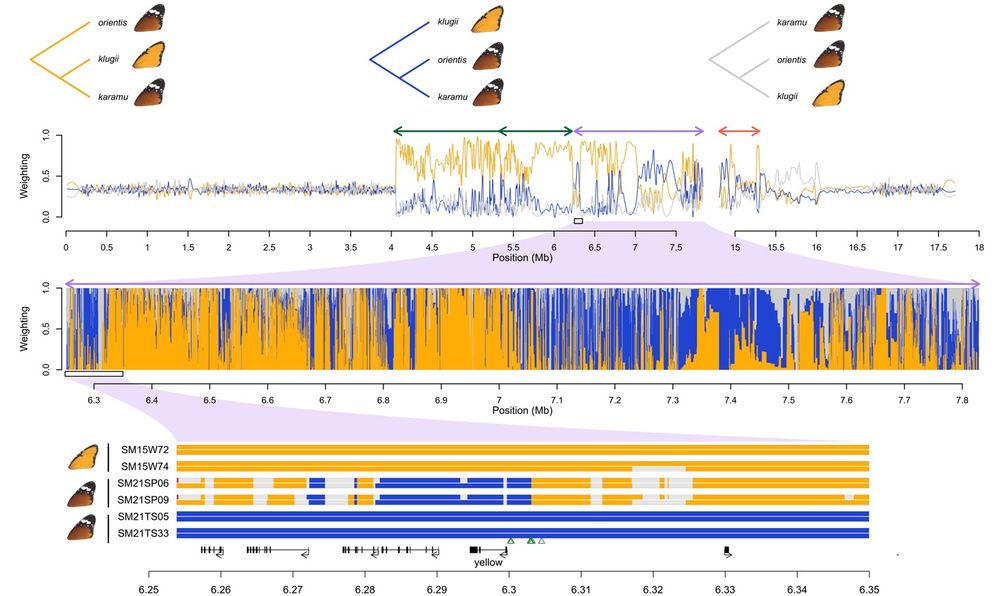
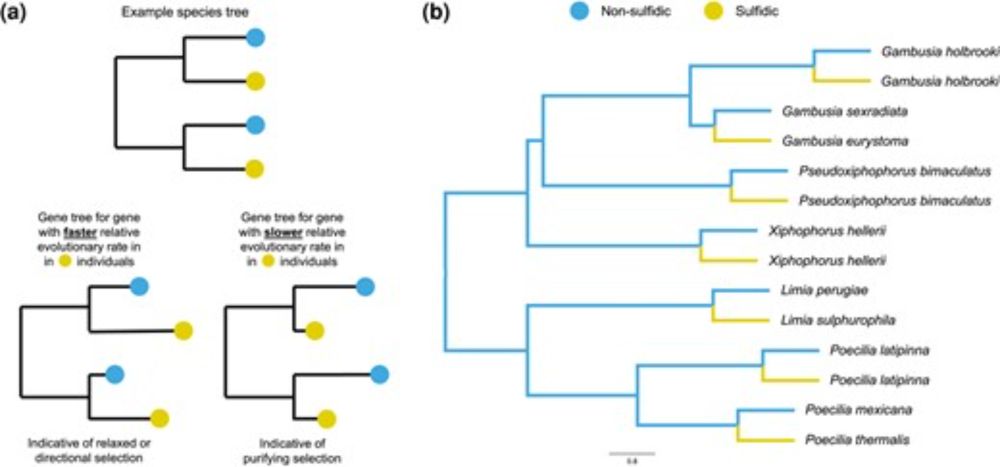
Top: Topology weightings across chr15 showing how the karamu haplotype is related to the klugii and orientis haplotypes. Upper panel shows three possible rooted genealogical topologies. Second panel shows weights for each topology along the chromosome, smoothed with a 20 kb span. Arrows above the plot indicate the locations of inversions. Third panel shows unsmoothed topology weightings across a 1.5 Mb region corresponding to Inversion 2. Bottom: Ancestry painting across a 100 kb region within Inversion 2, showing ancestry tracts for two homozygous karamu individuals compared to two representative individuals homozygous for the orientis and klugii haplotypes. Coding regions are indicated below the plot, with the candidate gene for background colouration yellow indicated. Green triangles represent the top 10 SNPs for background colour in our GWAS. There is evidence for recombination throughout the supergene region, and specifically in the vicinity of yellow, consistent with the hypothesis that orientis ancestry at this locus (i.e., the B allele) is associated with darker colouration in karamu individuals.
Dynamics of a supergene. A study of the BC supergene in wing color morphs of the African monarch #butterfly by @rishidekayne.bsky.social &co reveals dynamic evolution of #supergene haplotypes, fueled by incomplete recombination suppression 🧪 @plosbiology.org plos.io/3DiFhnL
03.03.2025 10:00 — 👍 53 🔁 31 💬 0 📌 2
Excited to share our review paper "Why Do Some Lineages Radiate While Others Do Not? Perspectives for Future Research on Adaptive Radiations" which is now online here: cshperspectives.cshlp.org/content/earl... Check it out for all things adaptive radiation! 🐟🦎🌿🦋
06.05.2024 14:40 — 👍 9 🔁 3 💬 0 📌 0
Hi everyone! It's about time I introduced myself - I'm Rishi and I'm doing a Postdoc at UC Santa Cruz working on adaptation genomics in Poeciliid fishes and Danaus butterflies. I'm particularly the role(s) of structural variation in evolution! 🧬 🧪
03.10.2023 17:25 — 👍 8 🔁 0 💬 0 📌 0
The Integration of Speciation network aims to facilitate discussion and promote knowledge exchange between all corners of speciation research, including through our virtual seminar series. Find out more: https://speciation-network.pages.ist.ac.at/
Postdoc @ University of Chicago Ecology & Evolution
Evolutionary genomics mostly
Postdoctoral researcher at Jagiellonian University, PhD at ETH Zürich/Eawag.
Interested in genetic variation, parasites and immune diversity. Likes most invertebrates and some of the vertebrates.
Based in New Zealand. Research: #seafood #genomics #adaptation. Editor-In-Chief for @EvolAppJournal. Prof at @AucklandUni. Science Group Leader @@bioeconomyscience
https://marenwellenreuther.com/
Collaborative research network on genome structural variants funded by the European Society of Evolutionary Biology (ESEB).
Immigration lawyer | Liberal | YouTuber | @SuperLawyer Award Winner | I sue USCIS and Consulates.
Senior Scientist of Coastal and Marine Ecology Division at GUIDE. Oceanography and Marine biologist.
Evolutionary geneticist with a particular interest in sexual dimorphism and sex chromosome evolution. Senior Lecturer at University of Sheffield www.alisonewright.co.uk
PhD Candidate, Laval University
Evolutionary Biology and Population Genomics applied to the study of Migration and Dispersal for the conservation of Salmonids 🐟🇨🇦
Ecology, conservation, statistics, reproducibility https://camargue.unibas.ch Retire statistical significance https://nature.com/articles/d41586-019-00857-9
Scientist. Group leader at the Sanger Institute, Cambridge UK. Somatic mutation and selection in normal tissues, cancer and ageing.
Assistant Professor @uarizona – Biological data science and macroevolution – Website: https://datadiversitylab.github.io/
Professor of EECS and Statistics at UC Berkeley. Mathematical and computational biologist.
Custom illustrations for biologists by Julie Johnson. • #sciart #scicomm • species paintings • scientific diagrams • figure design for ecologists and evolutionary biologists • www.lifesciencestudios.com
Scientist. Co-founder/CEO @ Arcadia Science. Board @ Astera and The Navigation Fund. Texan in CA.
Associate Professor in Plant Evolutionary Genomics 🌱🌸🌳🌼🌵
adaptation | repeated evolution | speciation | Arctic-alpine plants
Natural History Museum, University of Oslo, Norway 🇳🇴🦕
🌐 https://siribirkeland.github.io/
Staff scientist in the Sudmant lab at UC Berkeley
Genomics initiative lead at @GoogleDeepMind.
Models from our team: Enformer, AlphaMissense, and AlphaGenome.
Assistant Professor at SASTRA University | Multidisciplinary enthusiast 💬 | #Bioinformatics #SystemsBiology #DataScience #AMR #Coding #AI