Run shell commands in a lightweight sandbox (using `bubblewrap`)
Run shell commands in a lightweight sandbox (using `bubblewrap`) - sbx.sh
Safeguard yourself: do backups (yes, now!), don't run random code on production systems. If you must run untrusted pipelines, you can bubblewrap them in a bwrap sandbox - here is an example (works transparently with conda envs & system tools): gist.github.com/vmikk/08212e...
10.11.2025 09:14 — 👍 2 🔁 0 💬 1 📌 0
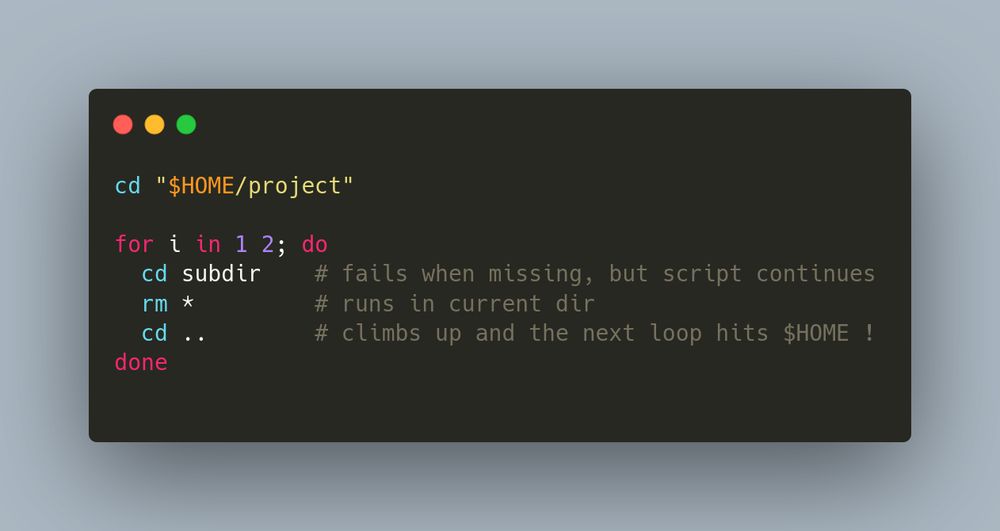
Terminal screenshot showing a minimal bash loop demo: starting in $HOME/project, a for loop runs cd subdir (which fails if missing), then rm * in the current directory, followed by cd .., so on the next iteration it climbs to $HOME and rm * risks deleting files there.
20+ years on #Linux, 🧬🖥️ #bioinformatics pipelines daily - and one relaxed moment running a colleague's script nearly wiped my $HOME 😅 Here's how a silent cd+rm combo escaped my project dir, the original script was quite tangled, but it hid this construct [ ⚠️ danger zone, don't run 🔥 ]
10.11.2025 09:14 — 👍 5 🔁 0 💬 1 📌 0

New fungal phylum-level system, with red font indicating novel taxa and size taxonomic diversity.
Fungal phylum-level classification update now available! Used "innovative taxonomy" to describe >100 taxa, from species to phylum, based on long reads, characteristic nucleotides and env-samples.
mycokeys.pensoft.net/article/1616...
#fungi #taxonomy #PacBio #classification #eDNA
20.10.2025 17:27 — 👍 24 🔁 13 💬 0 📌 1

New article on equitable reuse of public sequencing data, published in @natmicrobiol.nature.com!
Led by the Data reuse core team @lhug.bsky.social @environmicrobio.bsky.social Cristina Moraru, @geomicrosoares.bsky.social, @folker.bsky.social and with Anke Heyer and The Data Reuse Consotrium!
26.09.2025 19:34 — 👍 35 🔁 20 💬 0 📌 4
Who lives underground?
Find out now in our new paper published in @nature.com.
Key finding: 90% of predicted mycorrhizal biodiversity hotspots lie outside protected areas.
Read here: buff.ly/WmDqAP3 🧵
23.07.2025 15:20 — 👍 38 🔁 27 💬 1 📌 3
Post-Doc position for 3 years available jointly between University of Tartu (Estonia) and University of Aarhus (Denmark) on soil microbiome and biogeography. Skills required: bioinformatics, metagenome analysis, microbiology
Contact: leho.tedersoo@ut.ee
23.01.2025 06:54 — 👍 54 🔁 38 💬 0 📌 0

A newly discovered fungal species emerges from the open pages of a scientific book, surrounded by various lab journals and books, with research tools scattered around. The image highlights the connection between scientific discovery and publishing.
🍄📖 Do high-impact journals lead to more fungal species discoveries? Think again! Low or even no-impact factor journals contribute significantly! Let's value research quality over impact metrics in funding and hiring decisions! Read more @MycoKeys: mycokeys.pensoft.net/article/136048
21.11.2024 11:05 — 👍 6 🔁 1 💬 0 📌 0
PostDoc at the IBE @ibe-warszawa.bsky.social
in FunDive project @fundive-fungi.bsky.social
Soil animal diversity. Who eats whom in soil and what does this mean. #SoilBON. @soilorganismsj.bsky.social journal. #GlobalCollembola
This is the official feed for the Fedora Project, a global free & open source software community sponsored by Red Hat.
We make Fedora Linux - an innovative free & open […]
🌉 bridged from ⁂ https://fosstodon.org/@fedora, follow @ap.brid.gy to interact
PhD|He/Him
- All submitted job offers are important and will immediate take
- Plant Microbial Computational Bioinformatics | Sequence based ML/DL using Rust 🦀
- I don’t use and hate 🚫 LLM 🚫 AI for code
- Post Typos are meaningless autocorrect
🚫 Politics
A British Mycological Society journal (@britmycolsoc.org.uk). We publish investigations into all aspects of fungal ecology.
Account/posts by Editors-in-Chief Benjamin Wolfe(@benwolfe.bsky.social) and Allison Walker(@fungaldreamteam.bsky.social).
a.k.a. boB Rudis • 🇺🇦 Pampa • Don't look at me…I do what he does—just slower. #rstats #js #duckdb #goavuncular•👨🍳•✝️• 💤• Varaforseti í Gögn Vísindi @ GreyNoise • 47-watch.com • https://stormwatch.ing • https://dailydrop.hrbrmstr.dev • Maine🦞
Plant community ecology | Botany | Vegetation science | Natural habitats | Macroecology | Biodiversity | Conservation | Global change | Scientific publishing || Professor @ Masaryk University, Brno, Czechia
Follow us on Mastodon!: https://floss.social/@kde
A worldwide technology community 🌍 Creators of the Plasma desktop & a variety of Free and open source applications that let you control your digital life.
Get more KDE news at https://floss.social/@kde
Senior Scientist, Section Head, Team Leader at EMBL-EBI. PI of MGnify and co-lead of Ensembl, passionate about microbes and microbiome research. Bit of a tech geek too.
Prof. at Centre for Microbiology and Environmental Systems Science at University of Vienna, Austria and at Aalborg University, Denmark. Director of Cluster of Excellence "Microbiomes drive Planetary Health", Dad, Views are my own.
Microbiologist, JMF UniWien
Scientific/Research Officer in the Joint Research Centre (JRC) - European Commission. Managing the EU Soil Observatory - Developing soil health models & policy advice in Land Degradation. Own views
UNITE is a database and sequence management environment centered on the eukaryotic nuclear ribosomal ITS region.
Marine biologist interested in conservation genomics, fish biodiversity and eDNA 🐟💧🧬
PostDoc at the Globe Institute, Copenhagen | #biodiversity | #invasions | #eDNA | #OceanOptimist | 🏔️ 🌊 ☕️ 👨💻 📈
Postdoctoral Scientist @HIFMB | biodiversity data interoperability
Understanding eukaryotic molecular diversity and evolution. Marie Skłodowska-Curie Postdoctoral Fellow @IBE_Barcelona @beaplab.bsky.social
https://miguelmsandin.github.io/
https://thelifeofretaria.github.io/
Mastodon: @SandinMM@ecoevo.social
Keen on fungal genetics, fungicide resistance, phenomics, genomics, image analysis, epigenetics, epistasis, pleiotropy, host-pathogen interaction, TEs.
We want to understand how pathogens adapt and cause diseases. Also passionate about TEs, pangenomes & population genomics, bioinformatics and conservation genomics.
Professor of Evolutionary Genetics @ University of Neuchatel 🇨🇭
Hobbies: 👧🏻👧🏼🏔️🏕️