Hi Chris. So it would have to be high resolution and DDA data. The main idea here was to use environmental sample types for which we have exact coordinates. While most data we have up to this point are from environmental materials such as soil, wild animals would sure be interesting to add as well!
27.08.2025 21:06 — 👍 0 🔁 0 💬 0 📌 0
Great, thank you! I assume the data are already on MassIVE then. Then we just have to make sure to get all relevant metadata into redu.gnps2.org/selection/
27.08.2025 03:33 — 👍 0 🔁 0 💬 1 📌 0
Thank you for your interest! I have sent you an email and also turned private messages on here on bluesky for others.
27.08.2025 00:20 — 👍 0 🔁 0 💬 0 📌 0
Yes reach out to Yasin for this project. It will allow a worldwide picture of the environmental metabolome. This can be already published or not yet published data but will need world coordinates.
26.08.2025 21:46 — 👍 10 🔁 5 💬 2 📌 0

Interested in a co-authorship?
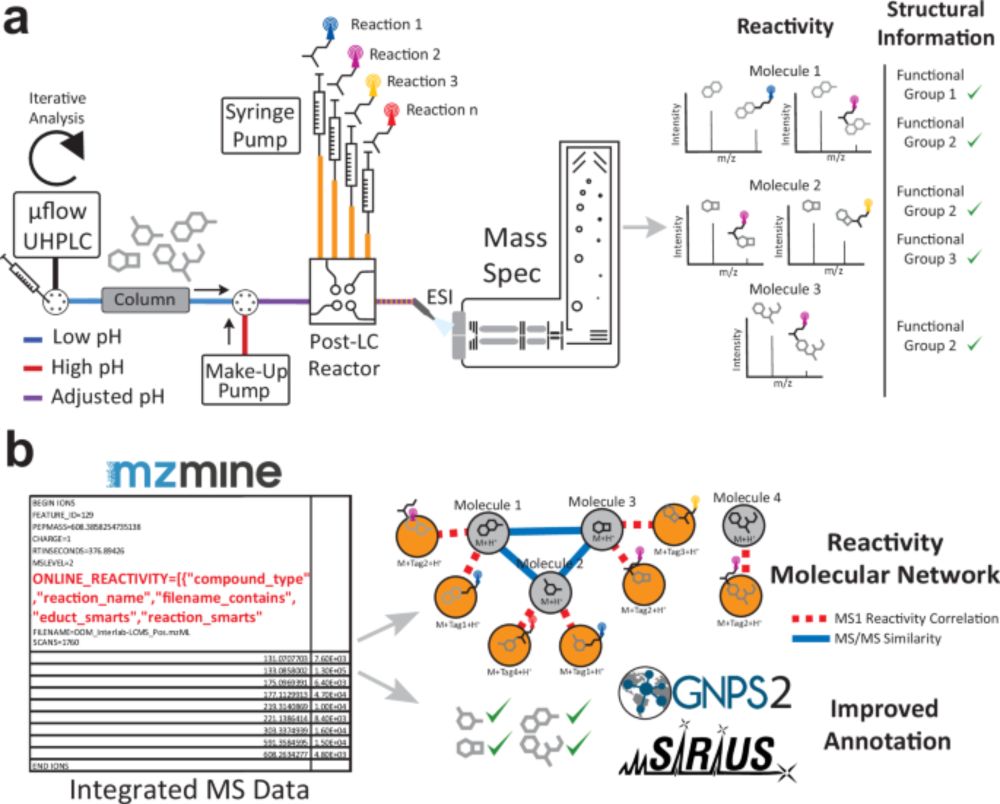
We’re building a tool for repository-scale untargeted #metabolomics and #exposomics of #environmental data. To make it the best it can be, we’re looking for people willing to share high-resolution LC-MS/MS (DDA) data from #water, #soil, #sediment, and related samples.
26.08.2025 19:51 — 👍 15 🔁 12 💬 3 📌 1

A Perspective on Unintentional Fragments and their Impact on the Dark Metabolome, Untargeted Profiling, Molecular Networking, Public Data, and Repository Scale Analysis.
In/post-source fragments (ISFs) arise during electrospray ionization or ion transfer in mass spectrometry when molecular bonds break, generating ions that can complicate data interpretation. Although ISFs have been recognized for decades, their contribution to untargeted metabolomics - particularly in the context of the so-called “dark matter” (unannotated MS or MS/MS spectra) and the “dark metabolome” (unannotated molecules) - remains unsettled. This ongoing debate reflects a central tension: while some caution against overinterpreting unidentified signals lacking biological evidence, others argue that dismissing them too quickly risks overlooking genuine molecular discoveries. These discussions also raise a deeper question: what exactly should be considered part of the metabolome? As metabolomics advances toward large-scale data mining and high-throughput computational analysis, resolving these conceptual and methodological ambiguities has become essential. In this perspective, we propose a refined definition of the “dark metabolome” and present a systematic overview of ISFs and related ion forms, including adducts and multimers. We examine their impact on metabolite annotation, experimental design, statistical analysis, computational workflows, and repository-scale data mining. Finally, we provide practical recommendations - including a set of dos and don’ts for researchers and reviewers - and discuss the broader implications of ISFs for how the field explores unknown molecular space. By embracing a more nuanced understanding of ISFs, metabolomics can achieve greater rigor, reduce misinterpretation, and unlock new opportunities for discovery.
If you’ve been following #metabolomics literature, you’ve probably seen a lot of debate on in-source fragmentation. We’ve put together a manuscript to clarify what it is, how to deal with it, and what it means for discovery in #metabolomics and #exposomics.
doi.org/10.26434/che...
23.08.2025 02:31 — 👍 4 🔁 3 💬 0 📌 0

The mass spectrometry of microbiome-mediated metabolism of food: challenges and opportunities
With the exception of molecules acquired through the lungs, skin absorption, or part of a medication regime, nearly all molecules in our bodies origin…
The interactions between food, microbiome and host that modulate health can be complex. Here, we offer a perspective on how mass spectrometry can be leveraged to address some of these challenges to understand host and microbial metabolism of food. A step closer to personalized health and nutrition.
08.08.2025 18:14 — 👍 4 🔁 3 💬 1 📌 0
GNPS2 and associated services will be down for power maintenance tonight and into tomorrow.
02.08.2025 04:48 — 👍 2 🔁 1 💬 0 📌 0
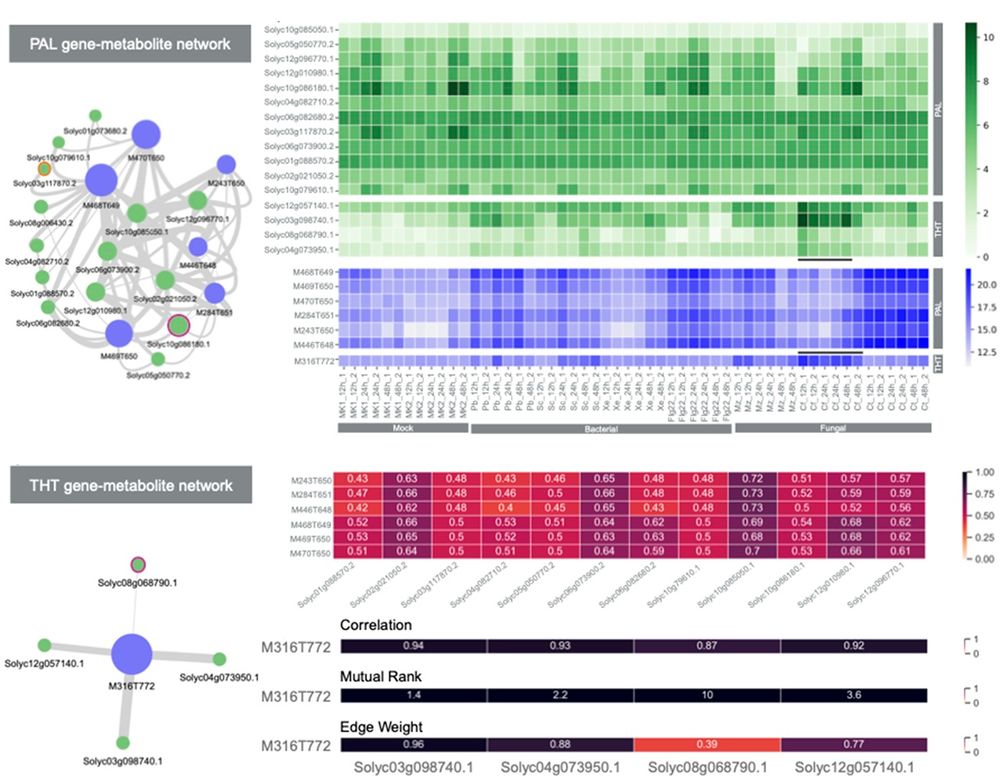
Detection of functional clusters (FCs) specific to the phenylalanine (PAL) and p-coumaroyltyramine (THT) pathways. Top left: Network depicting the relationship between transcripts and mass signatures within the PAL FC. Bottom left: Network illustrating the interplay between transcripts and mass signatures within the THT FC. Top right: Heatmap illustrating the expression levels of all transcripts within the PAL and THT FCs. Upper middle right: Heatmap displaying the abundance of all mass signatures present in the PAL and THT FCs. Lower middle right: Correlation matrix highlighting the correlations among transcripts and mass signatures within the PAL FC. Bottom right: Correlation matrix displaying the relationships between transcripts and mass signatures within the THT FC, including Mutual rank and transformed edge weights.
Elucidating #plant #Biosynthetic pathways: @jjjvanderhooft.bsky.social @marnixmedema.bsky.social &co develop #MEANtools, an unsupervised computational workflow that integrates #MultiOmics data to predict #metabolic pathways by linking transcripts to metabolites @plosbiology.org 🧪 plos.io/4odL94g
31.07.2025 08:49 — 👍 8 🔁 8 💬 0 📌 0
Excellent news: 𝐒𝐞𝐛𝐚𝐬𝐭𝐢𝐚𝐧 𝐰𝐢𝐥𝐥 𝐫𝐞𝐜𝐞𝐢𝐯𝐞 𝐚𝐧 #𝐄𝐑𝐂 𝐀𝐝𝐯𝐚𝐧𝐜𝐞𝐝 𝐆𝐫𝐚𝐧𝐭!
𝐁𝐢𝐧𝐝𝐢𝐧𝐠𝐒𝐡𝐚𝐝𝐨𝐰𝐬 will develop ML models to predict whether some query molecule has a particular bioactivity or is binding to a certain protein, where the only information we have about the query molecule is its tandem mass spectrum.
28.07.2025 11:16 — 👍 5 🔁 1 💬 1 📌 0
🚀 We’ve launched the new MassBank! Now live at massbank.eu & massbank.jp — redesigned with a faster backend, better search, and powerful tools for exploring & sharing mass spectral data. Enjoy the fresh experience! Feedback and ideas welcome, please post them on github.com/MassBank/Mas...
16.07.2025 18:57 — 👍 14 🔁 5 💬 0 📌 0
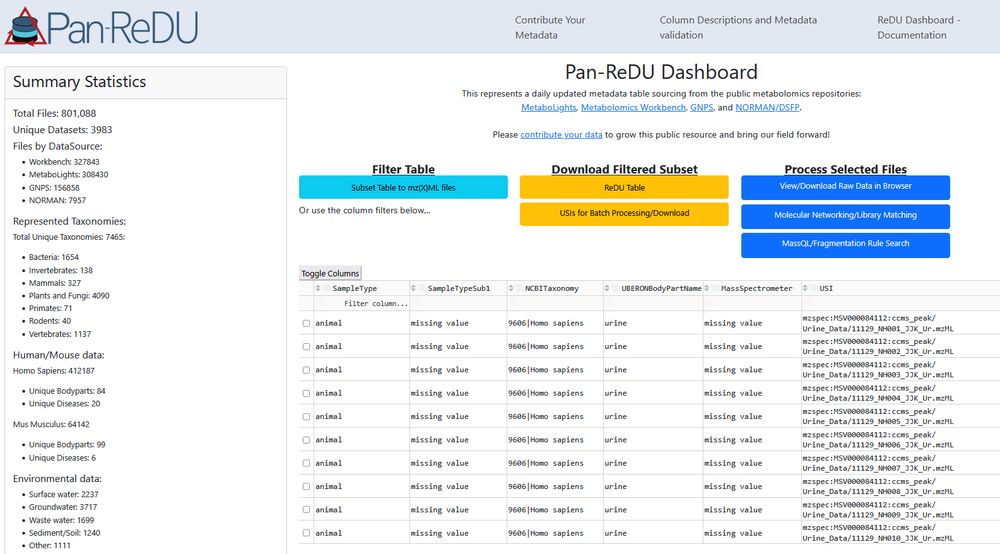
We just crossed the 800,000 files mark in Pan-ReDU. That's 800,000 public #metabolomics raw data files with harmonized metadata that can be re-analyzed to learn about new molecules and bio-distributions. 🎉 redu.gnps2.org
09.07.2025 00:55 — 👍 16 🔁 6 💬 0 📌 0
what is a good European and/or Open alternative to Feedly? I like something that works well on a phone as well as the web
Ideally, with CMLRSS support :)
27.06.2025 08:34 — 👍 1 🔁 2 💬 1 📌 0

@metabolights.bsky.social will be in #Metabolomics2025 in Prague!
Visit Posters 3007 C, 3006 C and 3000 C, say hi 👋 & discuss:
💻The most recent MetaboLights developments
🌐 Metabolomics Hub – a global open data consortium 🔗#ELIXIR Implementation Study on ontologies & semantic interoperability
19.06.2025 11:58 — 👍 14 🔁 6 💬 0 📌 1

Poster session at #ASMS2025 was as busy as always. Was great to see people agreeing that much remains to be discovered in untargeted #metabolomics. Thank you to all coauthors of the poster and associated paper. www.nature.com/articles/s42...
04.06.2025 19:39 — 👍 22 🔁 2 💬 0 📌 1
It’s so nice that an important paper led by yasin - is out. www.nature.com/articles/s41.... This paper is a key milestone as it is the foundation for data science across data repositories through indexing and metadata harmonization of 1.6 million files (although much more now due to updates). 1/n
30.05.2025 14:16 — 👍 20 🔁 7 💬 2 📌 0
Hope it is helpful. Also happy to answer questions!
28.05.2025 00:21 — 👍 1 🔁 0 💬 0 📌 0
This paper represents a great effort by @roman-bushuiev.bsky.social and his brother @anton-bushuiev.bsky.social. The DreaMS foundation model for mass spectra of small molecules now opens lots of avenues for possible downstream applications. It might be a game changer for computational metabolomics.
24.05.2025 08:21 — 👍 52 🔁 20 💬 0 📌 1

The Mass Spectrometry Query Language (MassQL) is an open-source language for instrument-independent searching across mass spectrometry data for complex patterns of interest via concise and expressive queries without the need for programming skills.
www.nature.com/articles/s41...
13.05.2025 15:25 — 👍 38 🔁 16 💬 1 📌 2
Senior Scientist
Senior Scientist
Please apply for this open position for a senior scientist at our faculty. It is the head of the mass spec center and you will be my close colleague! Application Deadline:8.5.2025
jobs.univie.ac.at/job/Senior-S...
30.04.2025 16:52 — 👍 8 🔁 9 💬 1 📌 3
Assistant Professor at the University of Toronto in Chemical Engineering and Applied Chemistry. Fascinated by marine microbes and their chemistry. 🌊, she/her, 🌈 https://www.gregorlab.com/
The official account of the Vardi Lab | Microbial Interactions in the Ocean | #phytoplankton #hostvirus #infochemical #stressresponse |
@WeizmannScience
Assistant Prof @Northeastern. Psychology + Biology. Neuroimaging, brain metabolism + mental health. Director of IASLab with Lisa Feldman Barrett & Karen Quigley
https://www.affective-science.org/
http://www.jordan-theriault.com/
Developmental and evolutionary ecologist studying marine invertebrates and their symbionts.
💼DOROTHY EU COFUND fellow
🔵Physoc Trustee
👂🏼Pronounced “Fair-on”
🌏Karolinksa Institute, Sweden
previously Maynooth Uni IRL, Uni of Galway IRL, QMUL London UK, Uni of Nottingham UK, TCD Dublin IRL
🥼obesity-assoc cancer immunology
❤️wild swimming
⚧️She/her
Microbial Ecologist 🦠
Research Fellow | Securing Antarctica's Environmental Future | Greening Lab | Monash University
Curious about the extremes of life 🏔️❄🌋🌊🏜️
they/them 🏳️🌈🏳️⚧️
linktr.ee/sophie.i.holland
SCIEX Mass Spectrometry Territory Manager - Eastern PA & DE
UD Alumni. Go Blue Hens!
MOBILion’s tech separates and analyzes challenging molecules with higher resolution, faster analysis, and simpler workflows—advancing safer, effective therapies
Physician-scientist leading a lab @lmumuenchen.bsky.social, visiting scientist
@broadinstitute.org | Writing about genetics, omics, deep phenotyping, precision medicine
https://www.deepvasc.com/
🇩🇪🇨🇦 Animal Metabolomics & Ecology Lab at Greifswald University. Chemical Ecology and Natural Products identification using LC-MS/MS and GC/MS to develop Urban Integrated Pest Management.
www.animal-metabolomics.com
Also husband and dad of my favorite 2
PhD bioengineering, BS civil engineering, human health, sustainability, data science, bioinformatics, metabolomics. Opinions are my own. (he/him)
Life scientist.
Head of EMBO Membership & Elections and Courses & Workshops @embo.org #EMBOevents
previously: Editor at Molecular Systems Biology, EMBO Press
Views my own
Mycologist and biotechnologist, Department Head at Helmholtz-Zentrum für Infektionsforschung, Acting President of the International Mycological Association and EIC of IMA Fungus
🏳️🌈Professor - Federal University of Uberlândia, Bioinorganic and (Bio)polymer Chemistry (views of my own).
(1) https://orcid.org/0000-0002-8357-3044
(2) https://www.linkedin.com/in/fernando-bergamini-7b577615/
(3) https://www.instagram.com/bioinspiredlab/
Prof. at Centre for Microbiology and Environmental Systems Science at University of Vienna, Austria and at Aalborg University, Denmark. Director of Cluster of Excellence "Microbiomes drive Planetary Health", Dad, Views are my own.
Development and application of efficient computational chemistry methods - based @unibonn.bsky.social.
This account is managed by group members of Prof. Stefan Grimme.
Professor of Natural Products Chemistry Université Paris-Saclay. Exobiology lover.