Join Prof. Matthias Mann, Ph.D. from the Max Planck Institute as he shares his first-hand experience with the SCIEX ZenoTOF 8600 system—a next-generation, high-sensitivity, compact, and quantitative…
Pushing proteomics forward: Prof. Matthias Mann on SCIEX ZenoTOF 8600 system at HUPO 2025
Watch Prof. Matthias Mann from the Max Planck Institute discuss the ZenoTOF 8600 at #HUPO2025. His presentation covers the transformative impact of this technology on protein analysis, focusing on enhanced sensitivity, quantitation, and real-world applications!
06.02.2026 14:54 — 👍 2 🔁 1 💬 0 📌 0

𝗪𝗵𝘆 𝗻𝗼𝘁 𝗷𝗼𝗶𝗻 𝗮 𝗻𝗲𝘄 𝗰𝗼𝗺𝗺𝘂𝗻𝗶𝘁𝘆 𝘁𝗵𝗶𝘀 𝘆𝗲𝗮𝗿❓ Check out the #BioACollective: Your Mass Spec Bioanalysis Community on #LinkedIn 👉 bit.ly/4kML0TU
#BioACommunity #Bioanalysis #MassSpectrometry #Biopharma #Pharma #SCIEX #MeetTheBioACommunity
02.02.2026 14:54 — 👍 0 🔁 1 💬 0 📌 0

Researcher Recess
🚨Philadelphia Researchers🚨
@sciex.bsky.social is hosting a Researcher Recess on 2/19 at White Dog Cafe in University City including:
-Delicious food & refreshing drinks
-Fun games and networking
-A chance to win exciting prizes
Save your spot here: sciex.com/Hidden/amer-...
#MassSpectrometry
02.02.2026 19:53 — 👍 0 🔁 1 💬 0 📌 0

𝗡𝗲𝘄 𝘆𝗲𝗮𝗿, 𝗻𝗲𝘄 𝗵𝗮𝗯𝗶𝘁𝘀? Join the #BioACollective: Your Mass Spec Bioanalysis Community to share ideas and meet like-minded people.
Join us on #LinkedIn today 👉 bit.ly/4kML0TU
#BioACommunity #Bioanalysis #MassSpectrometry #Biopharma #Pharma #SCIEX #MeetTheBioACommunity
20.01.2026 16:03 — 👍 0 🔁 1 💬 0 📌 0

SCIEX summit 2025 - a live virtual event
Hear from industry experts about the latest advancements in mass spectrometry
🧪 Dan Selle from Texas A&M reveals how the Echo® MS+ system helps identify covalent inhibitors in tuberculosis research.
Don’t miss this session at #SCIEXSummit2025
🔗 sciex.com/events/sciex-summit
08.10.2025 14:45 — 👍 0 🔁 1 💬 0 📌 0

SCIEX summit 2025 - a live virtual event
Hear from industry experts about the latest advancements in mass spectrometry
🧬 Patrick Schindler from #Novartis dives into IgA O-glycosylation using EAD and middle-up MS—advancing glycoprotein analysis.
Join live at #SCIEXSummit2025
🔗 sciex.com/events/sciex-summit
08.10.2025 15:25 — 👍 1 🔁 1 💬 1 📌 0
Interested in unraveling omega fatty acid positions in lipids? In this cooperation with @uni-grat.at, @ucsandiego.bsky.social we are able to use RP-MS/MS and LC=CL to achieve that. My lab contributed the EAD measurements using the ZenoTOF 7600 and the paper was published in @nature.com communication
12.08.2025 19:36 — 👍 2 🔁 1 💬 0 📌 0

A great showcase of @sciex.bsky.social's ZenoTOF 8600 system at #MSACL2025 today. Over 268,000 precursor and 12,000+ protein group IDs from just 200 ng of sample, with the Aurora Ultimate 25x75 XS & using Zeno SWATH DIA on the ZenoTOF 8600.
@davidcolquhoun.bsky.social, Ihor Batruch, & Patrick Pribil
25.09.2025 02:01 — 👍 5 🔁 2 💬 0 📌 0

Evaluation of the False Discovery Rate in Library-Free Search by DIA-NN Using In Vitro Human Proteome
Recently, deep-learning-based in silico spectral libraries have gained increasing attention. Several data-independent acquisition (DIA) software tools have integrated this feature, known as a library-free search, thereby making DIA analysis more accessible. However, controlling the false discovery rate (FDR) is challenging owing to the vast amount of peptide information in in silico libraries. In this study, we introduced a stringent method to evaluate FDR control using DIA software. Recombinant proteins were synthesized from full-length human cDNA libraries and analyzed by using liquid chromatography–mass spectrometry and DIA software. The results were compared with known protein sequences to calculate the FDR. Notably, we compared the identification performance of DIA-NN versions 1.8.1, 1.9.2, and 2.1.0. Versions 1.9.2 and 2.10 identified more peptides than version 1.8.1, and versions 1.9.2 and 2.1.0 used a more conservative identification approach, thus significantly improving the FDR control. Across the synthesized recombinant protein mixtures, the average FDR at the precursor level was 0.538% for version 1.8.1, 0.389% for version 1.9.2, and 0.385% for version 2.1.0; at the protein level, the FDRs were 2.85%, 1.81%, and 1.81%, respectively. Collectively, our data set provides valuable insights for comparing FDR controls across DIA software and aiding bioinformaticians in enhancing their tools.
Assessing FDR in proteomics using full proteome recombinant libraries: pubs.acs.org/doi/full/10..... A very elegant work and a benchmarking dataset useful for many other things too. Protein FDR higher than precursor FDR likely due to paralogues - and this needs to be researched further.
21.07.2025 08:29 — 👍 16 🔁 3 💬 2 📌 0
I'm really impressed by the Sciex developments with the ZenoToF 8600. Technically, it is 12k protein groups from 200 ng. It is a 3 species mixture. ~8k protein IDs are already a fantastic result.
18.06.2025 02:12 — 👍 1 🔁 1 💬 0 📌 0
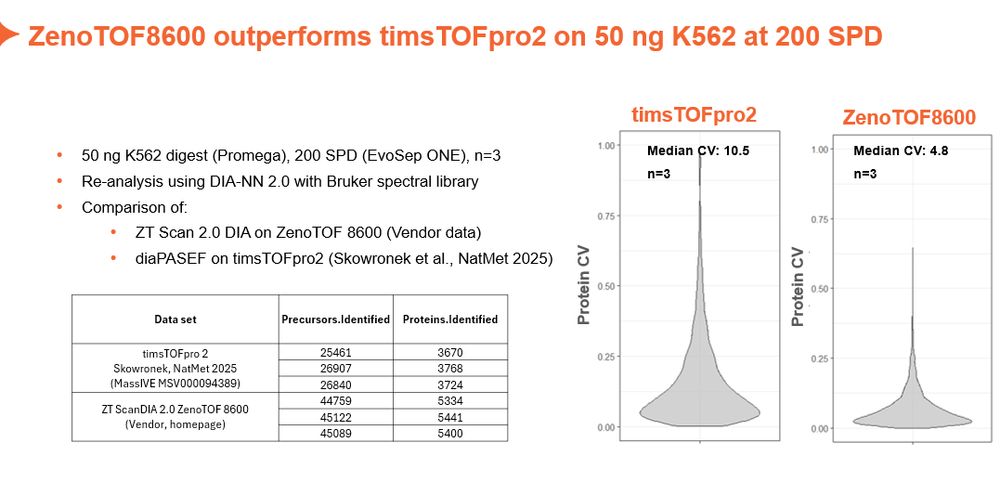
Sciex in the game.
Pretty impressed by ScanningSWATH data on the new ZenoTOF 8600.
#TeamMassSpec
27.06.2025 10:13 — 👍 11 🔁 2 💬 1 📌 0

Exciting showcase of the accuracy, speed, & sensitivity of the new ZenoTOF 8600 system at #EuPA2025 today.
268,000+ precursor & 12,000+ protein group IDs from 200 ng of sample using Aurora Series columns & Zeno SWATH DIA on the ZenoTOF 8600. Impressive numbers from SCIEX’s new flagship instrument.
18.06.2025 01:45 — 👍 9 🔁 2 💬 1 📌 0

#InTheNews: SCIEX Sets a New Standard in Accurate Mass Quantitation With the ZenoTOF 8600 System and New Software Collaborations 👏 🎉
🔍 Read the article 👉 buff.ly/tyywuRu
#DanaherNews #ZenoTOF8600 #SkepticsWelcome
07.06.2025 17:23 — 👍 5 🔁 2 💬 0 📌 0
📢 Attention #ASMS2025 attendees!!
Stop by the service and support booth in the SCIEX Hospitality Suite to get helpful answers from the people who know your instrument best.
Our expert team shares real-world advice from supporting thousands of instruments worldwide!
29.05.2025 17:28 — 👍 0 🔁 1 💬 1 📌 0

TODAY is the LAST DAY to register for the SCIEX 5K Spring / Stroll for Science, taking place Sunday June 1st from 8:30-11:30am at the Baltimore Harbor Promenade and the Maryland Science Center!
Throw your trainers and your running gear in your suitcase and register now 👉 sciex.li/1pw179 #ASMS2025
29.05.2025 18:00 — 👍 0 🔁 1 💬 0 📌 0

Something is coming... 👀
Get a sneak peek now 🔗 youtu.be/vnOJN_rAakw
#ASMS2025 #SkepticsWelcome
22.05.2025 12:02 — 👍 4 🔁 2 💬 0 📌 0
We are super thrilled to see ZT Scan DIA leading to insight from miniscule proteome samples, in clinical proteomics and functional proteomics studies that depend on highly precise protein quantification from large sample series.
14.05.2025 08:35 — 👍 2 🔁 1 💬 1 📌 0

Research on GLP-1 is crucial due to its widening therapeutic scope. #LCMSanalysis is used in #GLP1 research due to its sensitivity, specificity, and versatility. Check out the #SpotlightOnSeries for more information: bit.ly/41sahev
#Quantitation #SCIEX
29.04.2025 14:05 — 👍 0 🔁 1 💬 1 📌 0
Awesome application!!
11.03.2025 19:57 — 👍 1 🔁 0 💬 0 📌 0

Arthropod Parasitiformes amber inclusions. (A) Holothyrida, Neothyridae. A scavenger mite found together with insect inclusions in the amber from which it was separated for analysis. The figures provide details of the general look of the piece, measurements, and critical details for identification. Insect inclusions and amber without inclusions derived from this amber piece were processed as controls. (B) Cornupalpatum tick species for which stereomicroscope and Scan Ct images show the damage of the body preventing an accurate determination to the species level.

Neothyridae gen. sp. Elongation factor 1-alpha (A0A2H4RIF7). Sequence and phylogenetic analysis of the protein identified with highest coverage (244/342 amino acids, 71.3%). Neothyridae form a separate clade within Holothyrida mites.
Thread 1/5: PREPRINT: A major milestone on #Paleoproteomics?
Sciex 6600 TripleTOF analyses 99-million-year-old arthropods 🕷️ in amber.
Study identifies proteins from #Cretaceous #Holothyrida mites & #Cornupalpatum ticks!
de la Fuente et al. (2025) www.researchsquare.com/article/rs-6...
07.03.2025 07:47 — 👍 30 🔁 6 💬 1 📌 4

Mass spectrometry is a tool to deliver quantitative biological insights. Join Birgit Schilling at the SCIEX lunch seminar on Tuesday February 25th to discover how her lab is using the ZenoTOF 7600+ system to innovate proteomics workflows. #USHUPOSponsor #SponsoredPost
08.02.2025 21:00 — 👍 7 🔁 2 💬 0 📌 0

When is CID not enough for your research? At SCIEX, we’ve found some novel applications of hybrid and electron-based fragmentation to better characterize PTMs, including phospho- and glycopeptides. How else can EAD help biological insights? #USHUPOSponsor #SponsoredPost
14.02.2025 20:00 — 👍 2 🔁 2 💬 0 📌 0
Latest updates and posts from the #Proteomics subreddit (R/proteomics). http://reddit.com/r/proteomics
Join the conversation!
MS-based proteomics at University of Copenhagen
Old person forgetting everything they ever knew about proteomics at speeds in excess of 270 Hz...
UCLA Department of Chemistry and Biochemistry. Better living through mass spectrometry.
Ph.D. candidate in Smolka Lab; Weill Institute for Cell and Molecular Biology at Cornell University.
Also Phospho-Spectra Misalignment Specialist; Office of Fragmentation Fiascos
R.U. Lemieux Chair of Carbohydrate Chemistry
Professor, Department of Chemsitry
University of Alberta
A mass spec proteomics user in the CAR-T therapeutic space
Refurbished Lab Equipment Worldwide Since 1979
📍Chicagoland, IL
www.ietltd.com
Professor of Cell & Regenerative Biology and Chemistry at UW-Madison, Director of Human Proteomics Program🌱 | Passionate about Top-Down Proteomics, Cardiac Systems Biology, and Precision Medicine 🔬 | Optimist who loves gardening 🌸 Views are my own.
🔶 PhD student @ Dept of analytical chemistry | Palacký University | Olomouc, CZ
🔶 instrumentation engineer
🛠️ hardware archeologist
🛠️ obsolete device innovator
#️⃣ #hardwarearcheology #ChemSky #microfluidics
💬 ENG / UA / CZ
📝 opinions are my own
Analytical Chemist, Mass spec of all flavors, I work in industry and occasionally dip into academic stuff
Currently Mass Spec SW Marketing, former TMO SW Product Manager. Background in LCMS method development for topdown and bottom up proteomics.
Mass Spectrometry enthusiast! Science first!
Learning from the mistakes of others who took my advice 🤐 | Exploring Proteostasis with Mass Spectrometry 🧑💻🥼 | Science & curiosity in constant motion 🛣️| 📍 Germany
Senior Scientist @ Thermo (San Jose)
Enthusiast scientist, analytical technology nerd, metabolomics lover
Lead scientist at Liège University
Proteins | Sugars | Yeast | Beer | Mass Spectrometry | Professor in Biochemistry UQ. CW chickens, fungi, cats, pups.
Organic chemist in natural products, specialized in lipid derivatives. In love with oxylipins, mass spectrometry and computational organic chemistry.
Postdoctoral Fellow at Stanford University specializing in #spatialproteomics, #microscopy, deep visual #proteomics, advanced #image & #massspectrometry analysis. Passionate about unraveling cellular complexity through cutting-edge research and innovation🔬
Passionate about using mass spectrometry for studying disease. Clinical mass spectrometry (bioactive peptides and proteins) in the day, average triathlete in my spare time!



























