bims: Biomed News
A new week, a new selection of papers in #PlacentalCellBio
@biomednews.bsky.social
biomed.news/bims-placeb/...
Take a look at this pick from the Jawerbaum's lab in Argentina: pubmed.ncbi.nlm.nih.gov/40976335/
Enjoy!
28.09.2025 19:30 — 👍 1 🔁 1 💬 0 📌 0
Check this out: This is what the human fallopian tube looks like as one travels through its different sections.
We have developed a new 3D single-cell tissue mapping workflow to automatically/accurately detect ovarian cancer precancerous lesions (STICs).
More here: www.biorxiv.org/content/10.1...
25.09.2025 15:20 — 👍 41 🔁 9 💬 0 📌 1
New week, new summary of pubs in the #PlacentalCellBio
@biomednews.bsky.social
biomed.news/bims-placeb/...
My personal fav: this study about placental plasticity to hypoxia pubmed.ncbi.nlm.nih.gov/40961454/
Enjoy!
22.09.2025 14:37 — 👍 1 🔁 1 💬 0 📌 0

Scientists decry NIH pledge to end some human fetal tissue research
Move could signal return to ban on such studies imposed by first Trump administration
NIH says it will not renew a handful of research grants that an advocacy group identified as involving human fetal tissue—a decision that is setting off alarm bells for some in the scientific community. https://scim.ag/3VOoJJY
16.09.2025 15:12 — 👍 26 🔁 13 💬 2 📌 2

🚀 We’re all set!
This Sunday marks the return of the Placental Biology Course – our first in-person for six years. 25 participants joining for lectures and practical sessions delivered by experts in the field of placental biology!
05.09.2025 13:43 — 👍 2 🔁 1 💬 0 📌 0
bims: Biomed News
Here are my pre-selected papers in @biomednews.bsky.social 's #PlacentalCellBio of the week 🧐
biomed.news/bims-placeb/...
Having participated in this call as a member of the Fetal Membrane Society Consortium, my selfish pick for the week is: pubmed.ncbi.nlm.nih.gov/40840313.
Cheers!
04.09.2025 22:43 — 👍 2 🔁 2 💬 0 📌 0
We hope next to dig deeper into the mechanistic aspects that can fully explain this phenotype, expand on early (development) and late (cancer, metabolic disease) trajectories, and interrogate key environmental stressors that could potentiate or inhibit the disruption in tissue-specific autophagy.
03.09.2025 16:55 — 👍 1 🔁 0 💬 0 📌 0
Now, we have confirmed this phenotype in mice, demonstrated fragmentation of mitochondria, ↑LC3B-II
& SQSTM1↑ puncta (incl. short isoforms) and Nrf2–NQO1 detox pathway upregulation. This led to hepatomegaly with disrupted cellular architecture but no signs of early cancer as seen in other ATG KOs.
03.09.2025 16:55 — 👍 2 🔁 0 💬 1 📌 0

Identification of organelle-specific #autophagy regulators from tandem #CRISPR screens. New study from Truc Losier, Karyn King, Maxime Rousseaux, and Ryan Russell (@russellbiolab.bsky.social) @uottawa.ca: rupress.org/jcb/article/...
#Biochemistry #CellSignaling
28.08.2025 13:45 — 👍 7 🔁 5 💬 0 📌 1
bims: Biomed News
Here are the curated lists of pubs in the #PlacentalCellBio
@biomednews.bsky.social report from the last two weeks!
biomed.news/bims-placeb/...
biomed.news/bims-placeb/...
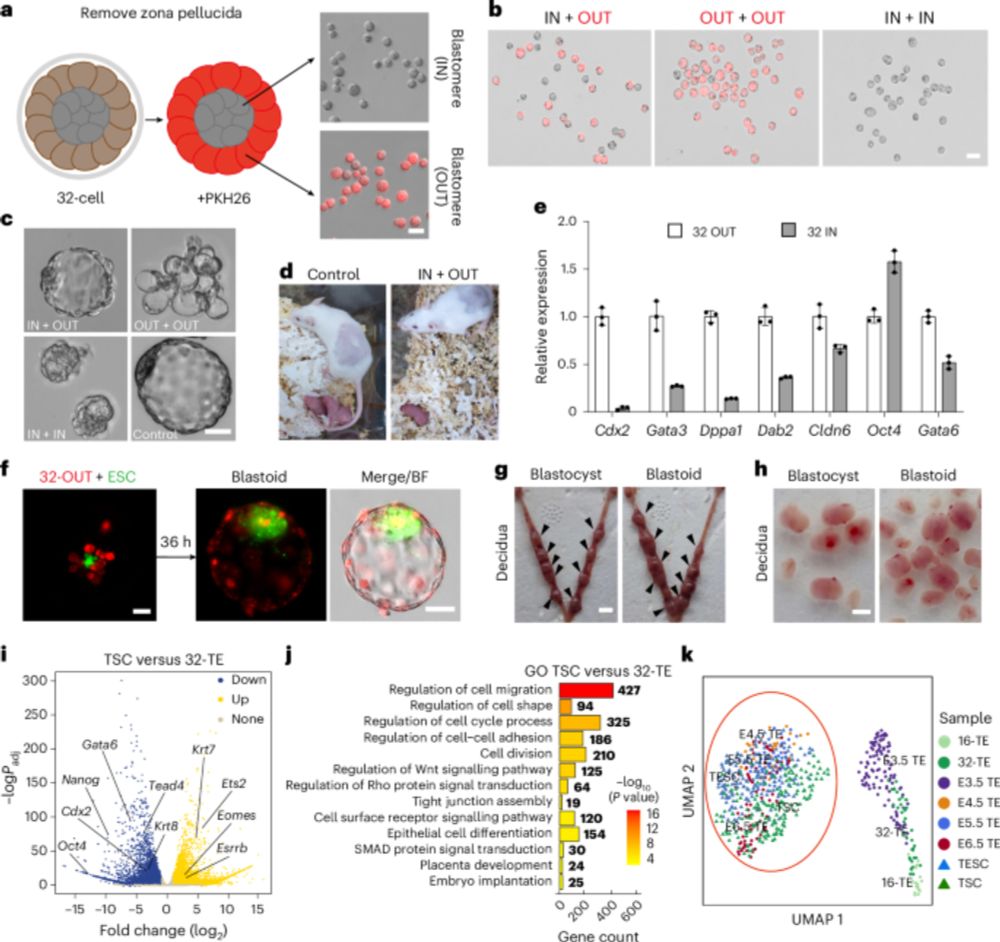
Lots of goodies but my personal choice: mTSCs from 32-cell morulae! www.nature.com/articles/s41...
Enjoy!
24.08.2025 17:01 — 👍 2 🔁 1 💬 0 📌 0

Dear Fly Community,
In May 2025, the NIH terminated all grant funding to Harvard University, including the NHGRI grant that supported FlyBase. This grant also funded FlyBase teams at Indiana University (IU) and the University of Cambridge (UK), and as a result, their subawards were also canceled.
The Cambridge team has secured support for one to two years through generous donations from the European fly community, emergency funding from the Wellcome Trust, and support from the University of Cambridge. At IU, funding has been secured for one year thanks to reserve funds from Thom Kaufman and a supplement from ORIP/NIH to the Bloomington Drosophila Stock Center (BDSC).
Unfortunately, the situation at Harvard is far more critical. Harvard University had supported FlyBase staff since May but recently denied a request for extended bridge funding. As a result, all eight employees (four full-time and four part-time) were abruptly laid off, with termination dates ranging from August to mid-October depending on their positions. In addition, our curator at the University of New Mexico will leave her position at the end of August. This decision came as a shock, and we are urgently pursuing all possible funding options.
To put the need into perspective: although FlyBase is free to use, it is not free to make. It takes large teams of people and millions of dollars a year to create FlyBase to support fly research (the last NHGRI grant supported us with more than 2 million USD per annum).
To help sustain FlyBase operations, we have been reaching out to you to ask for your support. We have set up a donation site in Cambridge, UK, to which European labs have and can continue to contribute, and a new donation site at IU to which labs in the US and the rest of the world can contribute. We urge researchers to work with their grant administrators to contribute to FlyBase via these sites if at all possible, as more of the money will go to FlyBase. However, we appreciate that some fu…

https://wiki.flybase.org/wiki/FlyBase:Contribute_to_FlyBase
Our immediate goals are:
1. To maintain core curation activities and keep the FlyBase website online
2. To complete integration with the Alliance of Genome Resources (The Alliance).
Integration with the Alliance is essential for FlyBase’s long-term sustainability. For nearly a decade, NHGRI/NIH has supported the unification of Model Organism Databases (MODs) into the Alliance, which we aim to achieve by 2028. Therefore, securing bridge funding to sustain FlyBase over the next three years is crucial for successful integration and the long-term access to FlyBase data.
At present, our remaining funds will allow us to keep the FlyBase website online for approximately one more year. Beyond that, its future is uncertain unless new funding is secured. We will, of course, continue pursuing additional grant opportunities as they arise.
Given the uncertainty of future NIH or alternative funding sources, we are relying on the Fly community for support. Your contributions will directly help us retain the staff needed to complete this transition and to secure ongoing fly data curation into the Alliance beyond 2028.
We at FlyBase are incredibly grateful for the outpouring of support from the community during this challenging time. Your encouragement has strengthened our resolve and underscores how vital this resource remains to Drosophila research worldwide.
Sincerely,
The FlyBase Team
The community of Drosophila researchers is amazing, mutually supportive and collaborative. Right now a key resource for our community, @flybase.bsky.social , is threatened by the cancellation of its NIH grant and is seeking community help in raising short term funds 1/n 🧪 please share
23.08.2025 12:18 — 👍 151 🔁 128 💬 1 📌 8

These are the most challenging times for early-career scientists and engineers.
My team is doing its bit to help by maintaining and even expanding our database of funding opportunities for early-career researchers.
We found 437 of them.
Download it freely here: research.jhu.edu/rdt/funding-...
11.08.2025 11:54 — 👍 312 🔁 200 💬 6 📌 3

The choice to have another pregnancy or not can be a difficult decision for #preeclampsia #eclampsia #HELLLPsyndrome survivors & their partners & is highly individualized. Speak with your patients during a preconception visit about what another pregnancy could look like for them
08.08.2025 15:49 — 👍 1 🔁 2 💬 0 📌 0
The transcription factor NR2F2 positively regulates the development of androgen-producing cells in the testes of fetal mice.
buff.ly/8SBhVjM
08.08.2025 08:21 — 👍 7 🔁 1 💬 1 📌 0
10 days left to apply for SDB #SciComm Internship. Deadline: August 15. www.sdbonline.org/science_comm...
05.08.2025 02:01 — 👍 7 🔁 6 💬 1 📌 0

Research Group Leader in Chemical/Synthetic Biology at MRC Laboratory of Molecular Biology
Start your UK & international job search for academic jobs, research jobs, science jobs and managerial jobs in leading universities and top...
We are looking to recruit a tenure track group leader in the field of Chemical/Synthetic Biology (in the broadest sense) to lead a research program within the Division of Protein & Nucleic Acid Chemistry ( www.jobs.ac.uk/job/DNW809/r... ) at the MRC Laboratory of Molecular Biology (LMB).
14.07.2025 12:09 — 👍 27 🔁 33 💬 1 📌 4
A fun reminder that a loss of 60,000 jobs in and around academia is ~50% more than all coal mining jobs in the entire country.
04.08.2025 00:02 — 👍 5376 🔁 1786 💬 51 📌 38

PIEZO1 drives trophoblast fusion and placental development - PubMed
PIEZO1, a mechanosensor in endothelial cells, plays a critical role in fetal vascular development during embryogenesis. However, its expression and function in placental trophoblasts remain unexplored...
After another fantastic @ssrepro.bsky.social annual meeting, here you have last week's selected paper in the #PlacentalCellBio @biomednews.bsky.social report:
biomed.news/bims-placeb/...
Highlight from our neighbors at Duke with an interesting role of PIEZO1 in STB formation!
Enjoy!
03.08.2025 15:45 — 👍 4 🔁 2 💬 0 📌 0
HEAL is a leading alliance of health and environment groups working at EU level. Working for better health, through a healthy environment.
📍 https://www.env-health.org/
✉️ press inquiries: nea@env-health.org
ℹ️: https://www.env-health.org/about/
Study genetic conflicts professionally. Try to avoid conflicts in personal life (with mixed results).
Fred Hutch Basic Sciences
(https://go.bsky.app/ReCeiC6)
UW Genome Sciences (go.bsky.app/L8RAbiJ)
HHMI (hhmi.bsky.social)
Posting in a personal capacity.
The American Physiological Society (APS) publishes 16 respected scientific journals that advance physiological research and communicate discoveries in physiology and medicine. Publish with Purpose (TM) in APS Publications. https://journals.physiology.org
#Leishmaniac, #FWO junior postdoc at @itmantwerp.bsky.social.
Using #singlecellgenomics to understand Aneuploidy and Genome instability in #Leishmania.
Professor, Oakland University, MICHIGAN.
Visiting Faculty@ LSI, UMich (Klionsky lab).
| Chromatin |Transcription |Autophagy | Yeast | Genomics |🧪
I'm into chord changes and cell biology.
Professor - University of Warwick. Director - The Company of Biologists. Views are my own and not those of any organisation I am associated with.
Mastodon: @steveroyle@biologists.social
Lab: https://roylelab.org
PhD student in Sabrina Büttner's lab
Lipid Droplets 💛 , Fluorescence Microscopy 🔬 and Proteostasis 🦠.
she/her
(Banner: Overuse of Unsharp Mask Function on confocal microscopy of yeast cells tagged with mCherry)
PhD student at Karolinska Institutet and Scilifelab | Immunobiophysics
https://www.csi-nano.org/
Developmental biologist, geneticist, & reproductive biologist. PhD. Son of an immigrant. Hispanic Filipino American. Endurance athlete.
Chemist in a CellBio place. Fan of lipids & membranes
Website: https://cocalc.com/
Collaborate in real-time via web browser while using your favorite programming languages and applications.
Like Google Suite for Computational Scientists.
Self-host your own instance of CoCalc: https://onprem.cocalc.com/
Solver of GTPases molecular puzzles | Membranes | CRISPR | STED super-res |Accidental immunologist | Academic mother of 2 cheeky monkeys | Assistant prof at FU Berlin. | Head of the Nanoscale Membrane Dynamics Lab
An enthusiast in cellular and organellar biology. Lives by seeing is believing.🔬
PhD student @Dartmouth
Studying cellular mechanisms of risk and resilience in neurodegenerative diseases. Views are my own.
The Triangle Cytoskeleton Meeting is an ASCB-supported local meeting in North Carolina's Research Triangle area.
Web development, data management and visualizations. Consulting services for scientific and r&d communities. https://www.researchelements.org/
Scientific content aggregator with custom content feeds. Science news, jobs, events and more. Sign up at https://www.scientific.today.
Postdoc Associate | interested in maternal-fetal tolerance & infection | #MHCIb #tunnelingnanotubes #TNTs #BaylorCollegeofMedicine #BCM
Biochemist and cell biologist working on autophagy at the Max Perutz Labs, University of Vienna