Great to see all these structures posted 😊 Amazing what endogenous extraction and purification can do. Congratulations to John and colleagues!
18.11.2025 20:59 — 👍 2 🔁 0 💬 0 📌 0

Cryo-EM maps and atomic models of the biotin-containing 3-methylcrotonyl-CoA carboxylase (MCC) complex and long-chain acyl-CoA carboxylase (LCC) complex
New lab preprint!
@zestytoast.bsky.social tagged a scarce mycobacterial protein in M. smegmatis with TwinStep but got… something? @kjamali.bsky.social's ModelAngelo built models & @martinsteinegger.bsky.social's FoldSeek IDed them as the biotin-containing MCC & LCC complexes
🧵
tinyurl.com/ukny4ptz
30.10.2025 03:21 — 👍 82 🔁 29 💬 3 📌 1
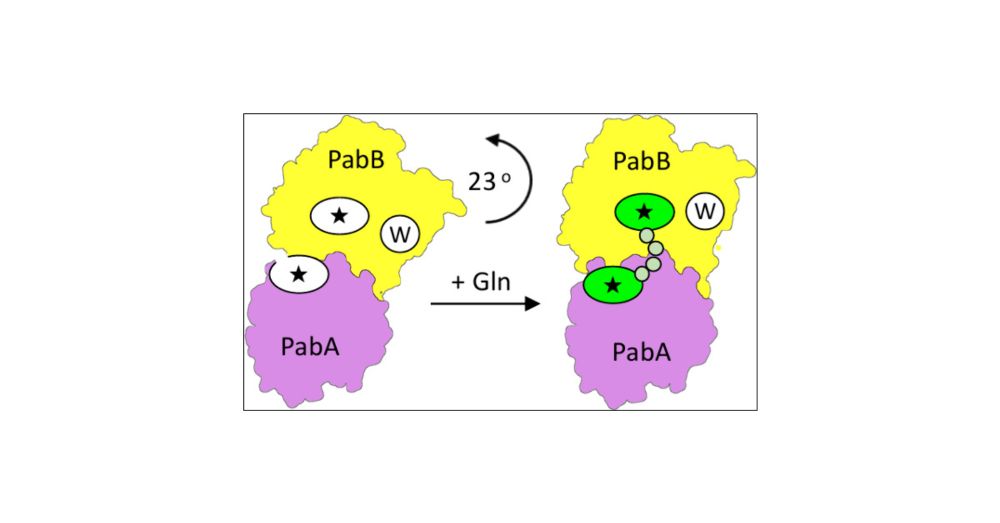
Binding of acyl-CoA substrates in AccA3/AccD4/AccD5/AccE5 causes a ~90 deg. rotation of the uneven 9-stranded (AccA3)4/AccE5 beta-barrel. The rotation propagates into the complete AccA3 assembly, which is split into two asymmetric AccA3 dimers.
18.11.2025 20:51 — 👍 4 🔁 0 💬 0 📌 0
A C-terminal helical dimerization module in the AccE5 subunit is responsible for the chimeric assembly of two related acyl-CoA carboxyl transferase subunits (AccD4, AccD5). Each of them specifically catalyzes carboxylation of long-chain and short chain acyl-CoAs, respectively.
18.11.2025 20:48 — 👍 2 🔁 0 💬 1 📌 0
Asymmetry, flexibility and sub-complex rotations of the AccA3/AccD4/AccD5/AccE5 holo complex are caused by the presence of a previously unknown epsilon-subunit (AccE5) in red/orange. AccA3, green shades; AccD4, purple shades; AccD5, blue/cyan shades.
18.11.2025 20:39 — 👍 2 🔁 0 💬 1 📌 0

Ensemble model of the full length Pex5 receptor (different blue color) in complex with Pex8 (green colors).
Key Findings:
Pex8 binds to a novel site on the mostly unfolded N-terminal region of the Pex5 receptor. This interaction is essential for peroxisomal protein import.
Computational modelling reveals the formation of an assembly with the trimeric peroxisomal E3-ubiquitin ligase.
We propose that this action positions the Pex5 receptor for its recycling, a step essential for the entire protein import process.
Why this matters:
Peroxisomes rely entirely on the import of folded proteins to function. Impaired peroxisome function is linked to severe disorders, and recent data show their crucial roles in carcinogenesis and the immune response. Our work elevates Pex8 from an enigma to a central player in this critical biological process.
Read the full story and see the structures here: https://www.biorxiv.org/content/10.1101/2025.08.30.673231v1
#CellBiology #StructuralBiology #Peroxisome #ProteinImport #bioRxiv #Biochemistry
We are excited to share our latest work, where we unravel the structural and functional secrets of the once-mysterious protein Pex8 revealing how it controls the peroxisomal cargo import receptor Pex5.
Read more here: www.biorxiv.org/content/10.1101/2025.08.30.673231v1
03.09.2025 10:28 — 👍 18 🔁 6 💬 1 📌 0
Professor of Chemistry @UFScripps
Interested in enzymology, bacterial metabolism, chem bio, mycobacteria, antibiotic discovery and antibiotic resistance. Former group leader at NIMR and The Crick.
"Dedicated scientist with deep expertise in Single Particle Cryo-EM, X-ray Crystallography, Protein Biochemistry, Molecular Biology, and Virology. Passionate about uncovering the complexities of biological systems and driving innovative discoveries."
Royal Society University Research Fellow | NUAcT Fellow @ Newcastle University @mhd-newcastle.bsky.social
Investigating mycobacterial transport systems
#structuralbiology #microbiology
Instagram @kshbeckham
The European Molecular Biology Laboratory drives visionary basic research and technology development in the life sciences. www.embl.org
Executive Director EMBL. I have an insatiable love of biology. Consultant to ONT and Cantata (Dovetail)
Computational Biologist, Head of research @ebi.embl.org, part-time
Heidelberg University, codirector DREAM challenges. For group's activities, see @saezlab.bsky.social
Statistician, Computational Biologist, R |> Bioconductor
https://www.huber.embl.de
Textbook: Modern Statistics for Modern Biology https://www.huber.embl.de/msmb/ (with @sherlockpholmes.bsky.social)
Pathogen informatics and modelling http://bacpop.org
(Research group leader at EMBL-EBI - http://ebi.ac.uk/research/lees/)
Head of Protein Sequence Resources at EMBL-EBI.
Group Leader at EMBL
Searching for PV = nRT in biology
The choreography of multicellular systems, computer simulations, gene regulatory circuits, and swarm robotics.
Head of EMBL Barcelona
Co-chair of the Barcelona Collaboratorium
Group Leader at EMBL Barcelona. Malaria, bioengineered vascular models, blood-brain barrier
Group Leader @EMBLBarcelona | PD @TheCleversLab | PhD @MITBiology (Jacks lab) | B.A. @NewCollegeofFL | She/her | 🇵🇷🇺🇸🇪🇺| #Organoids, #Neuroendocrine cells
Physicist trying to understand the behaviour of living organisms. Group leader at EMBL Barcelona.
Computational biology lab focusing on microbiomes in the human gut and the environment
Homepage: https://www.bork.embl.de/
Mastodon: https://mstdn.science/@BorkLab
Group Leader at EMBL Heidelberg. temperature, single cell genomics, proteins, and lots of fish.
The Prevedel lab @EMBL Heidelberg. We develop high-speed, deep tissue as well as Brillouin microscopy techniques. More details @ www.prevedel.embl.de
Biologist studying evolution, development, and gene-regulation. Group leader at EMBL, Heidelberg. Climber.
Scientist, Group Leader @EMBL Heidelberg.
Group Leader at EMBL spying on the social life of marine microbes and pushing for more inclusive sciences | Tara Oceans | Plankton Manifesto
https://www.embl.org/groups/vincent/
https://planktonmanifesto.org/