
I'm hiring a computational biologist interested in complex trait genetics using deep learning approaches. Reach out to me, if interested.
12.09.2025 19:00 — 👍 38 🔁 46 💬 1 📌 0@hakyim.bsky.social
Statistician doing genomic data science, faculty the University of Chicago, Korean, Argentinean, American. Love kimchi, math, science, books with beautiful prose.

I'm hiring a computational biologist interested in complex trait genetics using deep learning approaches. Reach out to me, if interested.
12.09.2025 19:00 — 👍 38 🔁 46 💬 1 📌 0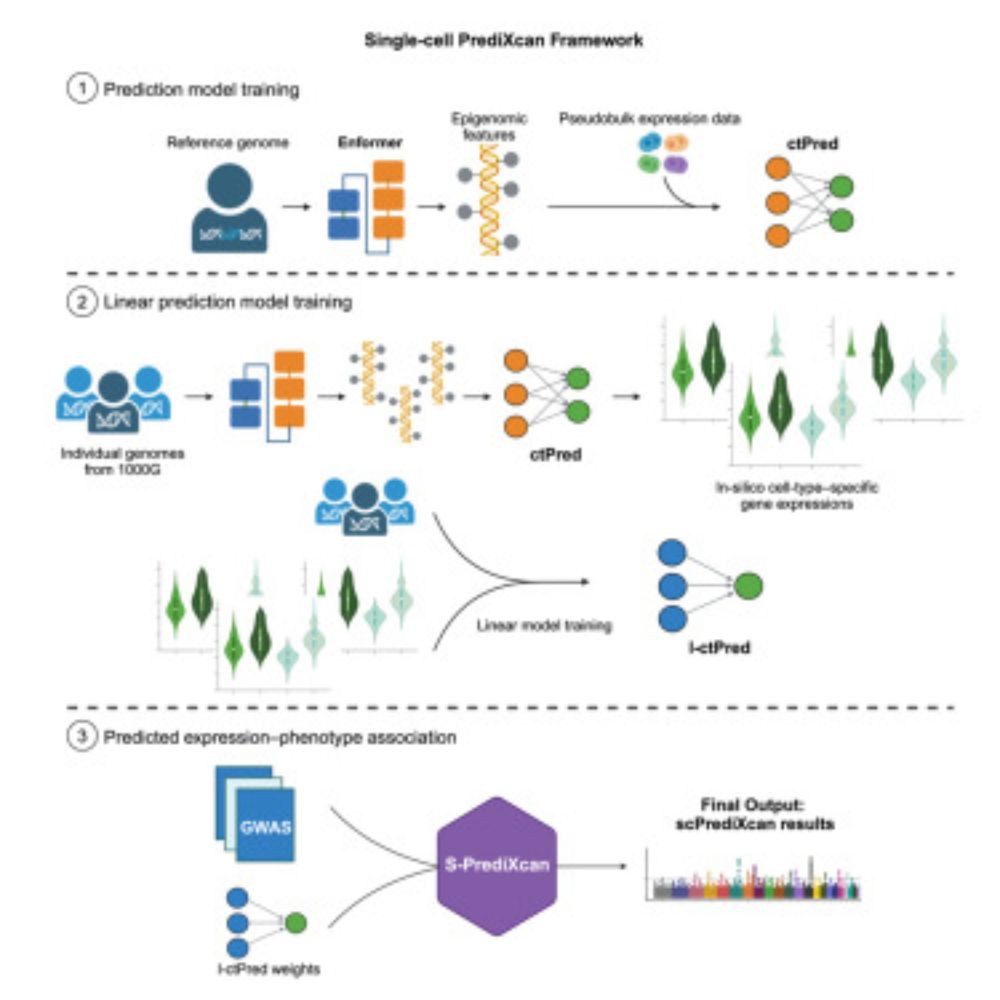
Check out our scPrediXcan paper
www.cell.com/cell-genomic...
Led by the talented @Charles_Zhou12 and supervised by @MengjieChen6
and me, with thanks to many contributors.
scPrediXcan integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework.

Need a break from doomscrolling?
Do you study the genetics of disease subtypes? Have you ever wondered how to efficiently run gene-based associations with multicategorical outcomes? Join us for the IGES Journal Club on Wednesday, February 26, 2025! 11AM ET.
Ughhh, suffering the plos system now. My collaborators won’t let me submit future papers to PLoS
11.02.2025 21:06 — 👍 1 🔁 0 💬 1 📌 0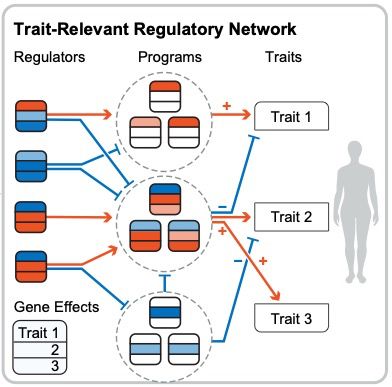
Modern GWAS can identify 1000s of significant hits but it can be hard to turn this into biological insight. What key cellular functions link genetic variation to disease?
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
1/At the #HoffmanLab lab meeting, we often have tech talks describing useful tools for other lab members. Since they might also prove useful for others, we've been posting almost every #HoffmanLabTechTalk for years. 🧪🧬💻
11.06.2024 21:24 — 👍 44 🔁 9 💬 2 📌 2with invaluable contributions from @temicrates Lisha Zhu @ssalazar_02 Sarah Sumner Hyunki Kim Saideep Gona @Festus_nyasimi Rohit Kulkarni @drjosephpowell @madduri
@boxiangliu

check out our new preprint led by Charles Zhou and supervised by Mengjie Chen and me doi.org/10.1101/2024... where we present scPrediXcan which integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework
15.11.2024 02:28 — 👍 29 🔁 13 💬 1 📌 0Consistency is key
13.11.2024 03:26 — 👍 1 🔁 0 💬 0 📌 0I’m at UChicago, develop methods to understand the biology of complex trait and diseases, aspire to help real people with my research. Currently very optimistic about predicting molecular traits from DNA sequences using m/billions of parameters
13.11.2024 03:25 — 👍 16 🔁 4 💬 0 📌 0🆕 Blog: Genomics scientists from and in Mexico community building lcolladotor.github.io/2024/11/11/g...
I want our voices🇲🇽 to be heard from the scientific community. But to be heard, we have to be a part of it
#LCG-UNAM #LCG-EJ-UNAM @cdsbmexico.bsky.social #NNB-UNAM #RMB
youtu.be/OVMw0k6AydA?...

Peter Buxtun, the whistleblower who revealed that the U.S. government allowed hundreds of Black men in rural Alabama to go untreated for syphilis in what became known as the Tuskegee study, has died. He was 86. apnews.com/article/buxt...
16.07.2024 15:18 — 👍 90 🔁 51 💬 2 📌 3IGES (international genetic epidemiology society) journal club recording "The All of Us Research Program: Data Resources and Access"; by Dr. Anji Musick is available here:
drive.google.com/file/d/1bty6...
Also worth noting: the US NIH offers financial support for those from ANY underrepresented group at ANY career stage to join an NIH funded lab!
Find a professor whose research you’re interested in and see if they are willing to host you! grants.nih.gov/grants/guide...
TWAS (transcriptome wide association study) is a statistical method that prioritizes genes that are more likely to cause a disease. It uses GWAS (genome wide association study) data, which is a method that identifies genomic loci associated with diseases
23.10.2023 14:53 — 👍 1 🔁 0 💬 0 📌 0Preprint is up now www.biorxiv.org/content/10.1...
21.10.2023 13:44 — 👍 4 🔁 3 💬 0 📌 1Preprint should be up shortly
18.10.2023 17:48 — 👍 2 🔁 0 💬 1 📌 04) LD is not necessary for the inflation to occur (our simulations were done using independent SNPs)
5) The inflation can be corrected by using the noncentral χ2 distribution with noncentrality parameter N h2δ Φ, where the factor Φ can be pre-calculated independent of the GWAS
2) Uncertainty in the prediction of the mediator does not cause inflation
3) Uncertainty in the prediction of the mediator reduces the power of the test
In summary
1) Polygenicity of the target trait induces inflation in the test statistics regardless of the genetic architecture of the mediating trait
Does this inflation affect other mediator-based *WAS?
Yes
What if we use PRS of GWAS traits to correlate with target traits? Is this going to be inflated?
Yes. You need to estimate Φ and use the noncentral χ2 distribution

Back to the effect of precision
Precision of prediction improves power, or equivalently prediction error reduces power but doesn’t increase inflation under the null

How well does your formula work under the alternative with finite N?
Pretty well

What happens under the alternative?
See figure for formula under the alternative
τ2 is the precision of the prediction

Can you estimate Φ?
Yes
See figure: most of the Φ are around 10e-5
Can you fix it?
Yes, just use the noncentral chi2 to compute the p-value instead of the standard chi2
pchisq(chi2, ncp=N*h2Y*phi, df=1, lower.tail=FALSE)
where phi=Φ is the gene specific factor
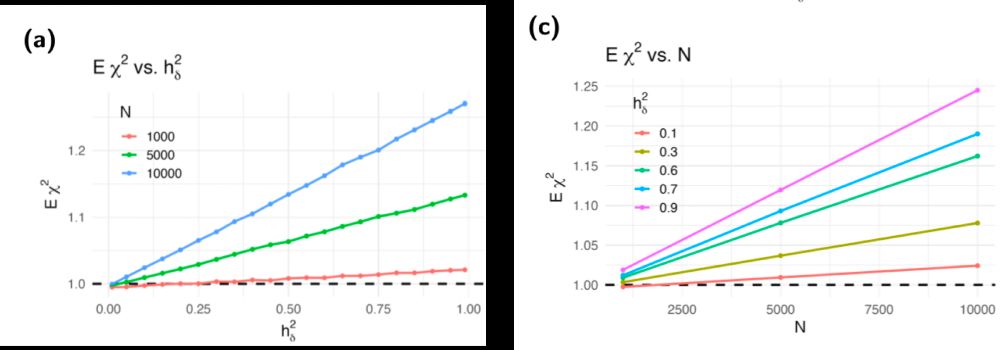
Does it matter for TWAS in practice?
Yes
See inflation in figure when using UK Biobank genotype data and null traits Y. It will continue growing as N grows.
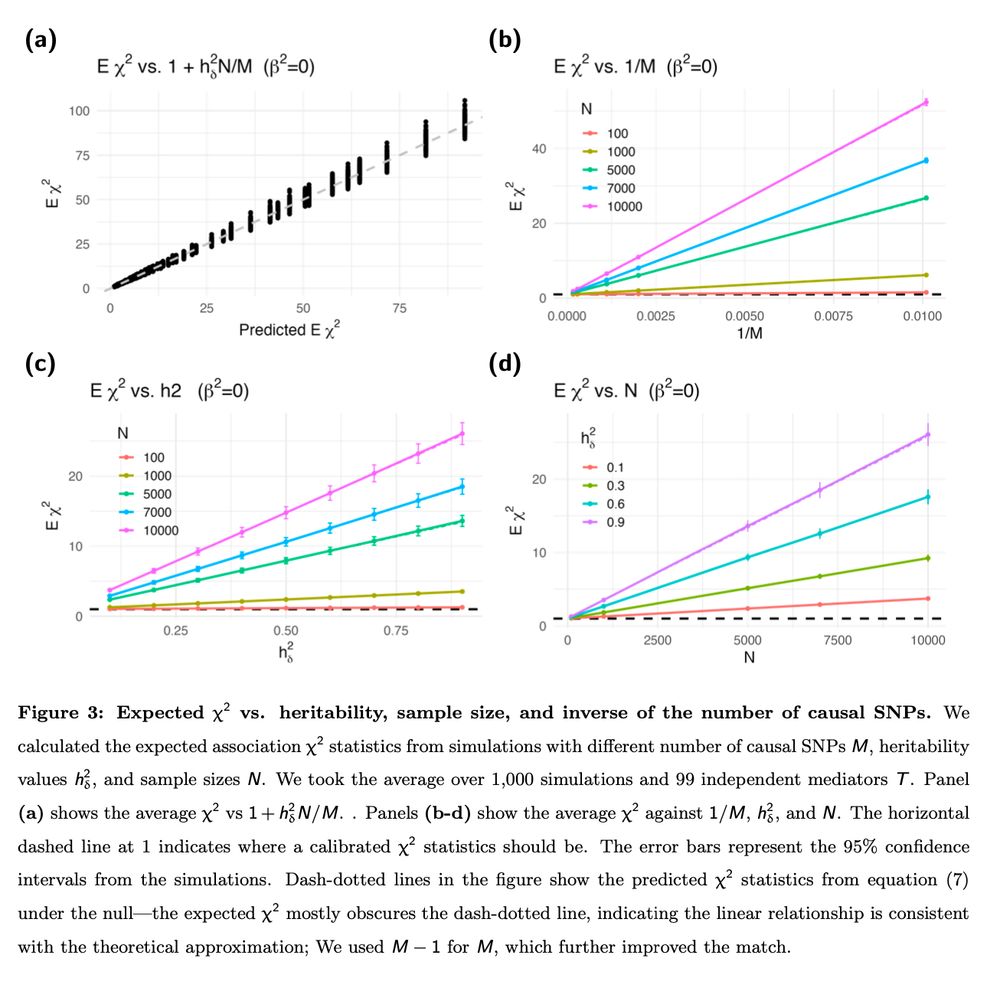
How we does your formula match with finite N?
Quite well
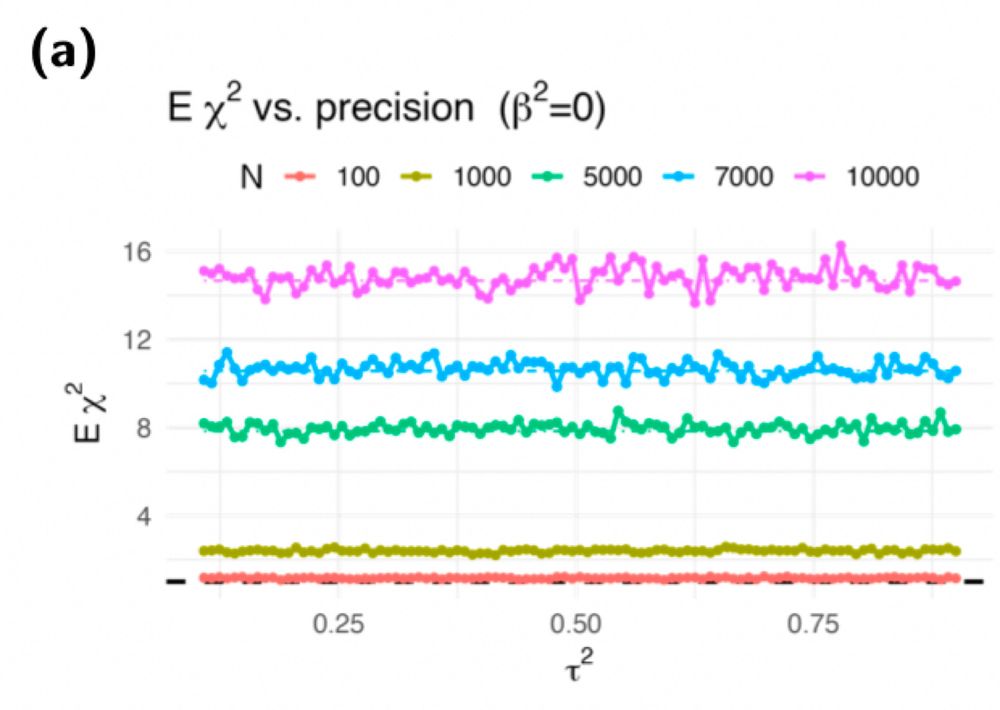
Does prediction error drive inflation?
No, it’s not the prediction error
See figure Eχ2 does not depend on the precision consistent with established error in variable literature
It’s the polygenicity of Y since Eχ2 = 1 + N h2 Φ. The inflation is 0 when the polygenic component of Y = 0
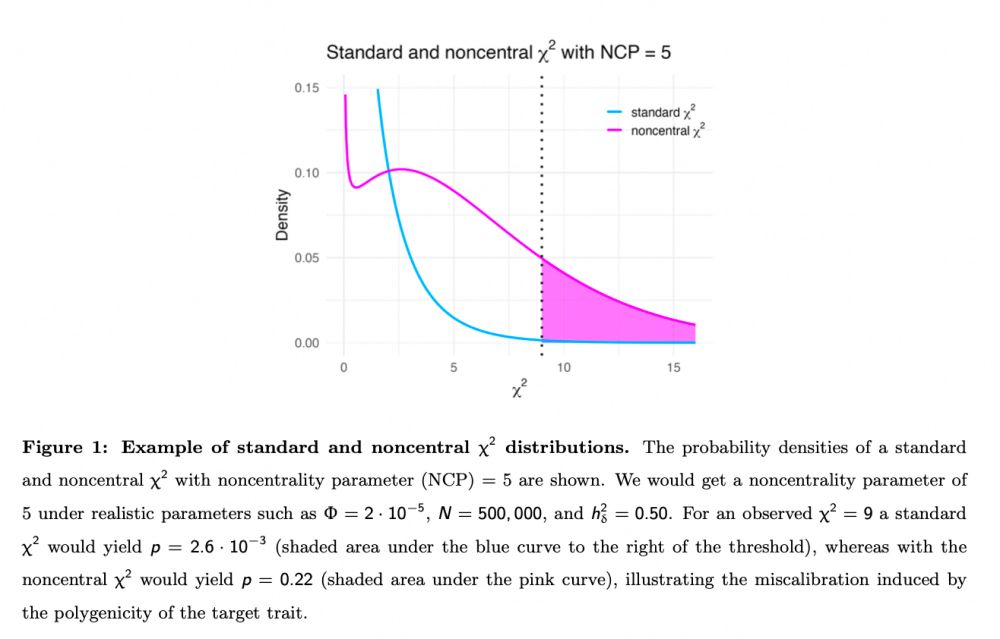
Does this matter?
Yes
See example in figure 1, pink area is much larger than the negligible blue area in this example