Especially relevant as a counterexample to OpenAI’s Prism tool, which will only worsen the science slop problem.
The point isn’t to be anti-AI. It’s to demand a better philosophy behind the tools we build
28.01.2026 21:25 — 👍 3 🔁 0 💬 0 📌 0
This tool is extremely helpful and I think all scientists could benefit from it. Good at all stages of the research process, especially the very beginning.
It's also a great of example of how we *should* be building AI tools for science. Not to replace scientific thinking, but to hone it.
28.01.2026 18:16 — 👍 12 🔁 4 💬 1 📌 0
lol I know someone like this. Works on their ‘Civil Liberties & Privacy Team’ (I know). Believes they can learn on the inside so they can regulate once they leave. Also, enjoys an upper class lifestyle
27.01.2026 00:51 — 👍 1 🔁 0 💬 1 📌 0
Interesting – this is what you need for synaptic plasticity to solve "within-neuron" credit assignment. Backpropagating APs, elevating Ca2+ above the plasticity threshold, should preferentially enter dendrites which recently activated.
Is this the first evidence of this?
22.01.2026 15:02 — 👍 2 🔁 0 💬 0 📌 0
What if there are many cells of each cell type? In that case I’d argue evolution is better suited to moulding populations. In cortex, genes don’t specify each neuron’s connectivity individually, nor could they, with far more synapses than bits in the genome
12.12.2025 15:23 — 👍 1 🔁 0 💬 1 📌 0
love it. somehow we’ve convinced ourselves that it’s only worth paying ‘the best and brightest’ to be curious for a living. It’s fundamentally an exclusionary vision for science when it ought to be inclusionary.
20.11.2025 00:23 — 👍 6 🔁 0 💬 0 📌 0
Instead of an ivory tower, science is shaped like a root system.
Like in a religion, local ‘churches’ provide lifelong learning for adults. Yearly calendar of holidays celebrating the miracles of evolution since the precambrian. Professors become pastors.
20.11.2025 00:17 — 👍 5 🔁 0 💬 1 📌 0
More of this type of content please!
20.11.2025 00:08 — 👍 0 🔁 0 💬 0 📌 0
The logic of 'rising stars' programs is that science needs to retain top talent. But if anything, I've seen more brilliant minds leave science due to the culture of individualist careerism that these awards contribute to, and are a symptom of.
12.11.2025 19:22 — 👍 19 🔁 3 💬 1 📌 0
For example, if you spatially cluster the brain based on transcriptomics (e.g. www.biorxiv.org/content/10.1...) the clusters barely match functional areas.
IMO, data converges: we need to revisit the atlas.
But I agree broadly: brain tissue is far from homogenous, and we neglect it at our peril.
06.11.2025 14:23 — 👍 2 🔁 0 💬 0 📌 0
It is possible to work on neural networks as a neuroscientist and actively oppose how AI is developing and how it will affect society.
02.11.2025 22:14 — 👍 11 🔁 0 💬 1 📌 0
My confidence in that last sentence was less than 100%. But what was it? And if I had said "absolutely positive" somewhere, what then?
If you can't quantify natural language uncertainty, you can't train for it. At least explicitly. Thus all we have are RLHF approaches
14.10.2025 21:00 — 👍 2 🔁 0 💬 1 📌 0
True, but I think that's something different. Among the uncertainties flying around in Bayes stats – epistemic uncertainty over model weights, aleatoric uncertainty reflected in output probabilities, etc – none of them capture hedging in natural language. (1/2)
14.10.2025 21:00 — 👍 1 🔁 0 💬 1 📌 0
One hurdle: how do you reliably quantify the uncertainty expressed in a natural language paragraph?
If you gave me this measure for all data in the wild, and it was differentiable wrt the model, one might take a likelihood-max approach (plus calibration etc)
14.10.2025 20:18 — 👍 1 🔁 0 💬 1 📌 0
OSF
Consciousness science as a marketplace of rationalizations
my commentary on @smfleming.bsky.social and @matthiasmichel.bsky.social's thought-provoking BBS paper, and more generally about the field.
osf.io/preprints/ps...
10.10.2025 18:05 — 👍 45 🔁 18 💬 2 📌 3
Truly. ‘The use of state power for personal vendettas and returning personal favors’ is what we see daily. It’s plain corruption and I don’t know why that’s not the front message
10.10.2025 03:55 — 👍 1 🔁 0 💬 0 📌 0
Truly crazy - Texas groups are demanding absolute ideological alignment.
There are multiple TT job calls at Texas public universities right now. I will not be applying.
19.09.2025 01:20 — 👍 2 🔁 0 💬 0 📌 0
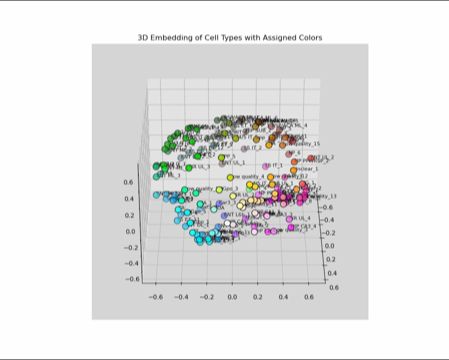
Under the hood, this
1) gets the avg (pseudobulk) gene expression of each type,
2) computes the similarity matrix,
3) projects expression into a 3D space using MDS, and
4) interprets the space as a perceptually uniform color space (LUV)
18.09.2025 16:03 — 👍 1 🔁 0 💬 0 📌 0
To make these colormaps, I made an easy python package:
```
!pip install colormycells
# Create a colormap based on cell type similarities
colors = get_colormap(adata, key="cell_type")
# Plot your cells
sc.pl.umap(adata, color="cell_type", palette=colors)
````
github.com/ZadorLaborat...
18.09.2025 16:03 — 👍 2 🔁 0 💬 1 📌 0
When colors reflect the actual differences between cell types, you can see things you wouldn't otherwise. For the mouse cortex, see how the layers form a rainbow, indicating a gradient of gene expression.
I also think this makes pretty plots 🌈 so that's nice.
18.09.2025 16:03 — 👍 1 🔁 0 💬 1 📌 0

Good scientific plotting uses color to convey meaning. We don't want to distract readers. Yet with default categorical maps,
(-) some cell types jump out for no reason
(-) perceptual similarity between colors is meaningless
(-) fewer colors than types, leading to repeats
18.09.2025 16:03 — 👍 1 🔁 0 💬 1 📌 0

Do you plot transcriptomic data, coloring each cell by its cell type? ~ Don't use a default colormap! ~
Instead, use colormaps that capture biological meaning. If two cell types are very similar, their colors should be similar too. Read on 🧵
🧬💻
18.09.2025 16:03 — 👍 9 🔁 2 💬 1 📌 0
This is real – Anthropic just agreed to a $1.5B class action settlement for authors of copyrighted works it stole. File your claim here: www.anthropiccopyrightsettlement.com
Unclear if this is just 'books' or journal articles too, but I'm putting mine in anyway
www.reuters.com/sustainabili...
08.09.2025 13:56 — 👍 2 🔁 0 💬 0 📌 0
I think this is the right view. But it’s worth adding that a large section of neuroscience defines specialization by what areas ‘encode for’, defined predictively.
Rather than the bell tolling for local specialization, it might be tolling for that ‘functional’ stim-response paradigm
04.09.2025 18:02 — 👍 3 🔁 1 💬 0 📌 0
what’s crazy is how they figured this out. makes for good science reading!
“Replication of an alien genome within one’s own cytoplasm echoes the endosymbiotic domestication of organelles [mitochondria]. Clonal males may thus be regarded as organelles at the superorganism level” 🤯
03.09.2025 23:44 — 👍 4 🔁 0 💬 0 📌 0
This interview of Monica López-Hidalgo by @analog-ashley.bsky.social is inspiring. All scientists’ job description should include this, great model
02.09.2025 20:50 — 👍 12 🔁 4 💬 0 📌 0
who is liable when an error is made by a BCI-using human with AI intervening in the output? The human? The BCI manufacturer? No one?
02.09.2025 12:16 — 👍 1 🔁 0 💬 0 📌 0
Get on my wavelength, man
30.08.2025 15:42 — 👍 4 🔁 1 💬 0 📌 0
Oh no how is it that "neuromodulation" means such different things across communities? Who let this happen?
For the record, the right answer is serotonin, etc.
The wrong answer is brain stimulation in a medical context, DBS, TMS etc. I will endlessly defend this turf.
29.08.2025 18:26 — 👍 1 🔁 0 💬 0 📌 0
PhD student at the Champalimaud Centre for the Unknown
https://asagodi.github.io/
Neuroscientist, Associate Professor, Lerner Lab. Dopamine and basal ganglia circuits controlling reinforcement learning and decision-making. Open/inclusive science. Happy working mom of 3.
Yale ➡️ UCSF ➡️ Stanford ➡️ Northwestern
lernerlab.org
Spinal cord and motor control scientist; PI at NINDS
Fighting every day to deliver a city that working New Yorkers can actually afford. Mayor of New York City.
AI and Games Researcher at NYU. Head of AI at Nof1.
PhD Student at MIT Brain and Cognitive Sciences studying Computational Neuroscience / ML. Prev Yale Neuro/Stats, Meta Neuromotor Interfaces
Bren Professor of Computational Biology @Caltech.edu. Blog at http://liorpachter.wordpress.com. Posts represent my views, not my employer's. #methodsmatter
NeuroAI PhD student @ Mila & Universite de Montreal w/ Prof. Matthew Perich.
Studying continual learning and adaptation in Brain and ANNs.
making computers better using brains and making brains better using computers. computational neuro and dumb little web apps
sticking a steam wand directly into my brain and frothing it like a cappuccino at @kordinglab. senior research scientist at JHU/APL
Bloomberg Distinguished Professor (SAIS & Carey Business School), Johns Hopkins University. Political economy, Brazil, and a little bit of futebol.
Neuroscientist and gardener
Lab site- saunderslab.com
Nerd, Husband, Dad, and Neuroscientist in Alabama. Flawed. Occasionally opines on football, cats, and beer. He/him
"Forth Eorlingas!"
https://hardawaylab.org/
Neuroscientist at CU Boulder. he/him. Mostly amplify other neuroscientists. www.root-lab.org
scientist.
tollkuhnlab.org
Diné & Chicano. Postdoctoral Fellow in the Maguire Lab at Tufts University. NIH-NIMH DSPAN K00. Arizona sports sufferer.
Brain scientist, tall human, dog owner in the twin cities
Neuroscientist, educator, author of SO YOU WANT TO BE A NEUROSCIENTIST? (Columbia, 2020) & MIND OVER MATTER (Princeton TBD) // professor at ucsd // co-host of www.changetechnically.fyi with @grimalkina // www.ashleyjuavinett.com
Professor of Behavioral Neuroscience at UCLA. Family-loving, vinyl-listening Angeleno 🫶🏼 she/her
Posts are my own and do not reflect my employer.
computational cognitive science @ nyu. director NYU minds, brains, and machines initiative. https://gureckislab.org. Are you interested in research in my lab? https://intake.gureckislab.org/interest/




