🧬🖥️ oarfish v0.9.0 is out! The big improvement in this version is future-oriented. v0.9.0 allows the user to pass in transcript sequences via the `--annotated` option, the `--novel` option, or both. The provenance will be separately tracked to that later 1/4
29.06.2025 23:39 — 👍 17 🔁 6 💬 2 📌 0

Release v0.13.0 · COMBINE-lab/piscem
Install piscem 0.13.0
Install prebuilt binaries via shell script
curl --proto '=https' --tlsv1.2 -LsSf https://github.com/COMBINE-lab/piscem/releases/download/v0.13.0/piscem-installer.sh | sh
Downl...
🖥️🧬 The new release (v0.13.0) of piscem is out. However, almost all changes here are transparent to the user (except for the need to rebuild the index before using the new version). I want to take a second to talk about something that often goes unappreciated. 1/x
github.com/COMBINE-lab/...
31.05.2025 19:16 — 👍 17 🔁 3 💬 1 📌 1
@mikelove.bsky.social it's been a pleasure and an insightful experience collaborating with you. Thank you for taking out the time to attend my defense.
13.03.2025 21:45 — 👍 1 🔁 0 💬 0 📌 0
Excited and nervous at the time!!! Looking forward to it.
13.03.2025 03:05 — 👍 8 🔁 1 💬 1 📌 0

Release v0.19.0 · COMBINE-lab/simpleaf
Install simpleaf 0.19.0
Install prebuilt binaries via shell script
curl --proto '=https' --tlsv1.2 -LsSf https://github.com/COMBINE-lab/simpleaf/releases/download/v0.19.0/simpleaf-installer.sh | sh...
We just released simpleaf v0.19.0 (github.com/COMBINE-lab/...). Make sure to pair it with piscem v0.12 for faster scATAC-seq processing. The other big feature & first explicit @scverse.bsky.social integration point is AnnData format output with the new `--anndata-out` flag!
25.01.2025 15:04 — 👍 11 🔁 4 💬 0 📌 0
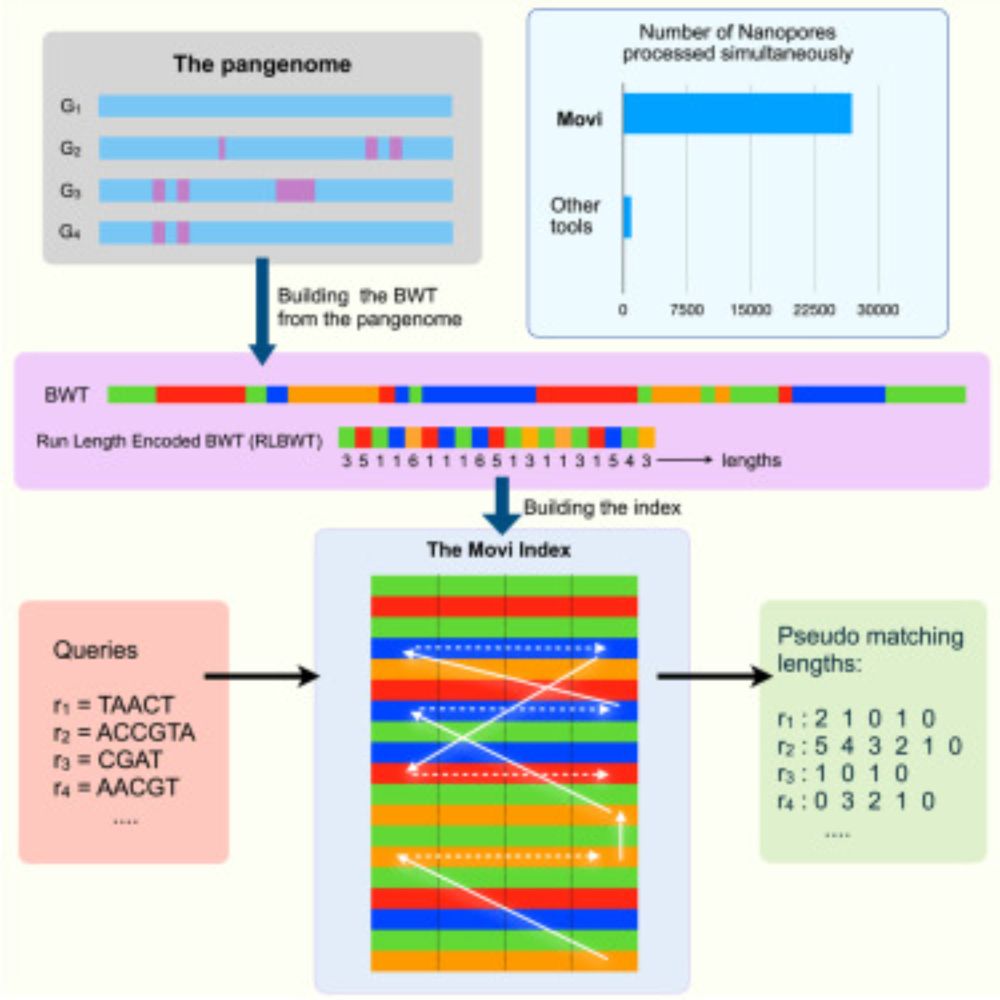
Movi: A fast and cache-efficient full-text pangenome index
Biocomputational method; Classification of bioinformatical subject; Genomic analysis
Very excited to see Movi (by @mohsenzakeri.bsky.social) now out in iScience: www.cell.com/iscience/ful.... Movi builds on the "move structure" pangenome index, a compressed full-text index and close cousin to r-index. Compared to r-index, the move structure is simpler and more cache-efficient.
11.12.2024 16:48 — 👍 44 🔁 17 💬 1 📌 0
It should definitely be doable if we store the rank and then we can perhaps later use it in the augmented bed file. We currently do support a SAM file as an output for the mappings for all the reads in the fastqs but then that does not do any deduplication/barcode correction.
05.12.2024 05:45 — 👍 2 🔁 0 💬 0 📌 0
PhD Student, Computational Biology
Machine learning, epigenomics, gene regulation, causal inference
University of Tübingen | University of Dundee
https://ntanmayee.github.io/
Senior Research Scientist @dfcidatascience @harvardchanschool
Cancer genomics, bioinformatics, chromatin & epigenetics, single-cell, plus occasional dog, food, and travel photos
AKA @jeremy_m_simon, @jeremy@genomic.social elsewhere
In Almouzni Lab at Institut Curie, a bioinformatician among H3 histone variants afficionados, exploring their role in shaping chromatin in pediatric brain cancers.
Dog dad. I work at PacBio.
https://www.unimedizin-mainz.de/medizinische-mikrobiologie-und-hygiene/
#DGfI #DGHM #IUIS / 🦖☄️🦠🔬🧫🐁🧍/ #immunometabolism / #host-pathogen interaction / 🇪🇺 / own views only
65 years ago, in a town that smells of rotten eggs & has no greenery of any kind, a premature baby boy was born to a teenage mother w/ recently broken bones, married to a sociopath who worked at a navy base 100's of miles from any water.
That baby was me.
Healthcare AI, Entrepreneur, Hardtech Mentor, Supply Chain Expert, Early Stage Startup Investor, ex-Toyota Motorola, Harvard Kennedy School
🏀 Dad, #StarTrek🖖🏽
Krishna Take The Wheel
CS PhD student at @jhu.edu @jhucompsci.bsky.social
Teaching machines to learn biology 🧬💻
https://khchao.com/
Associate Professor @ UC-Davis | Former Senior Director @ Exai Bio | Computational Biology, ML/AI, Genomics, RNA, Cancer
Genetic, research, and software engineer looking to make friends.
Creator of Poly, an open source Go package for engineering DNA 🧬
Let's engineer weird little guys together.
github.com/bebop/poly
📍San Francisco
Postdoctoral researcher, NCMM Oslo | TCR-seq, microRNAs, transcription factor binding sites, CRISPRi, colorectal cancer, immunotherapy, computational biology and bioinformatics.
PhD Student @ JHU Langmead Lab
Professor of Computer Science @ JHU. https://www.langmead-lab.org/ https://www.youtube.com/BenLangmead
Immunology PhD student @UMBaltimore. Views are my own. Favorite rabbit holes include #flowcytometry #rstats #bioconductor, with dashes of #coffee #kayaking #birding #ultrunning #languages. RPCV Ghana 16->18, TCK Mexico 96->11 https://github.com/DavidRach/
Associate professor @liigh-unam.bsky.social. Interested on theoretical population genetics and developing new computational methods to understand our past. He/him/his.
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Guest Scientist IMP Vienna, Board of Directors NumFOCUS
Incoming Prof UMass Chan Medical
Previously Stanford Genetics, UW CSE.
‖ Permanent Researcher / INRIA Start. Faculty @ INRIA Rennes 🇫🇷 ‖
BioInfo/CompBio: algorithms, genomics, pathogens & rapid diagnostic of antibiotic resistance《 https://brinda.eu | https://github.com/karel-brinda 》
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, Compute Acceleration in biotech // http://albertvilella.substack.com
Developing data intensive computational methods • PI @ Seoul National University 🇰🇷 • #FirstGen • he/him • Hauptschüler
The 29th International Conference on Research in Computational Molecular Biology. recomb.org
April 26 - 29, 2025



