
Looking for a postdoc in computational chemistry, machine learning, and enhanced sampling simulations?
The NAWA Ulam 2026 call is open -- reach out to me at jr@fizyka.umk.pl if interested!
nawa.gov.pl/en/nawa/news...
@jkrd.bsky.social
Associate professor at NCU, Poland. Developing advanced methods for atomistic simulations.

Looking for a postdoc in computational chemistry, machine learning, and enhanced sampling simulations?
The NAWA Ulam 2026 call is open -- reach out to me at jr@fizyka.umk.pl if interested!
nawa.gov.pl/en/nawa/news...

📢 PET-MAD is here! 📢 It has been for a while for those who read the #arXiv, but now you get it preciously 💸 typeset by @natcomms.nature.com Take home: unconstrained architecture + good train set choices give you fast, accurate and stable universal MLIP that just works™️ www.nature.com/articles/s41...
28.11.2025 08:36 — 👍 15 🔁 6 💬 0 📌 2
Great collaboration with Yanbin Wang and @MingChe40113998 from Purdue Uni. -- JCTC just published our paper introducing generalized sample transition probabilities (GSTP) for constructing collective variables from biased MD data.
pubs.acs.org/doi/full/10....
Now available in Journal of Chemical Information and Modeling!
pubs.acs.org/doi/10.1021/...

Together with my students, we have implemented a PyTorch Lightning package for dimensionality reduction with a parametric version of tSNE. More methods soon!
Paper: arxiv.org/abs/2505.16476
Code: github.com/NeuralTSNE

The paper describing our community effort to collect and organize #plumed tutorials has been published in the Journal of Chemical Physics, as part of the Michele Parrinello Festschrift! doi.org/10.1063/5.02...
04.03.2025 14:13 — 👍 31 🔁 11 💬 0 📌 1For those seeking postdoc opportunities in Poland -- NAWA has opened the Ulam program. If you are interested in applying for a scholarship in our group, please contact me!
nawa.gov.pl/en/programy-...
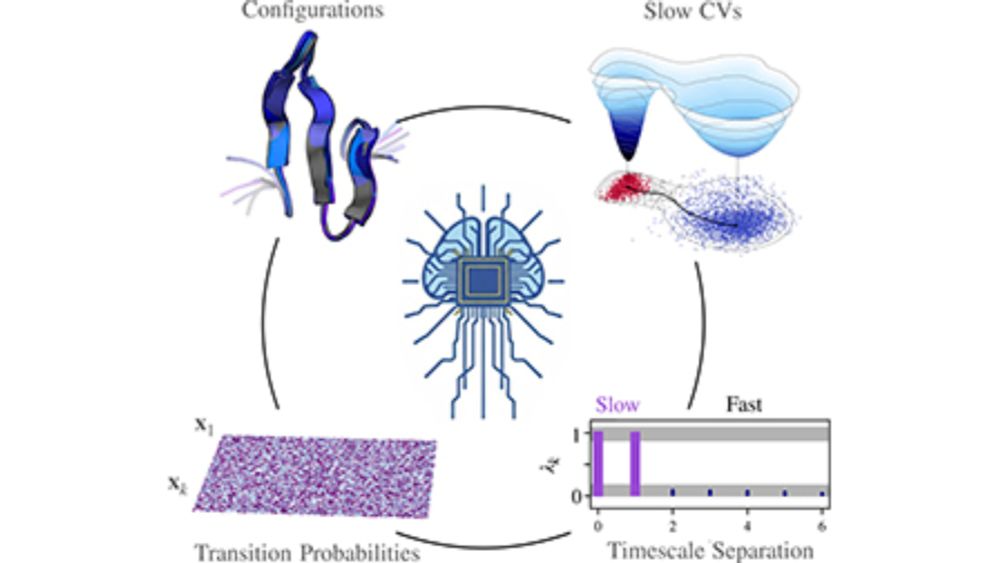
Very happy to announce that our featured review on thermodynamics-informed learning of slow CVs has finally been published in Chemical Physics Reviews as part of the Special Collection on AI and Machine Learning in Chemical and Materials Science!
doi.org/10.1063/5.02...

Our review on ML, CVs, and enhanced sampling has been accepted in Chem. Phys. Rev. Congratulations to Tugce Gokdemir
on her 1st first-author paper! arxiv.org/abs/2412.20868
Many thanks to Haochuan Chen, Luke Evans, Luigi Bonati, and Omar Valsson for their great feedback!
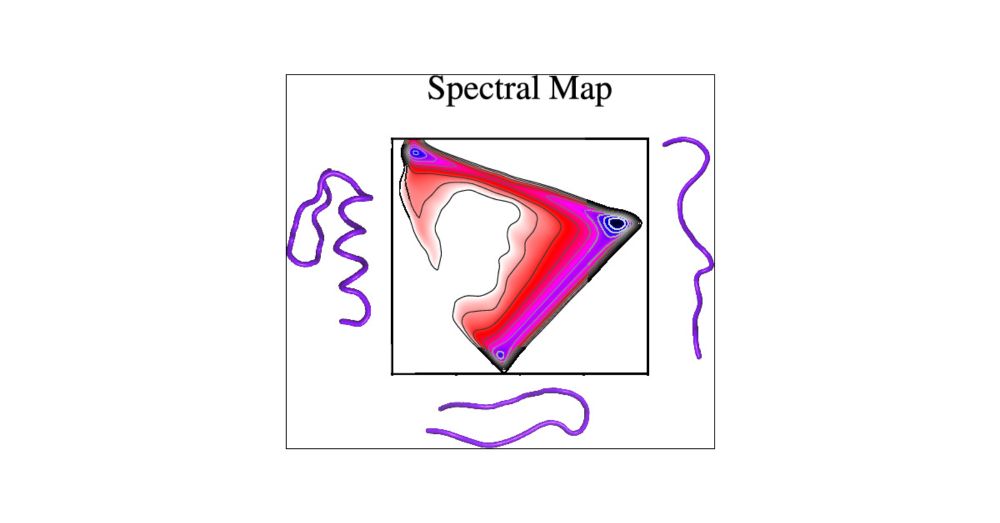
For anyone interested in spectral maps.
I made a typo in one equation in the original paper published in JCPL. This has now been fixed; see the accompanying correction note.
pubs.acs.org/doi/full/10....
Interested in doing MD simulations of protein-ligand dissociation? Check out our PLUMED tutorial on the new version of maze: www.plumed-tutorials.org/lessons/24/0...
Implementation: github.com/jakryd/plume...
More details about PLUMED Tutorials in our collaborative work: arxiv.org/abs/2412.03595

📢 New #preprint describing our community effort to share #plumed tutorials arxiv.org/abs/2412.03595 ! Explore the tutorials at www.plumed-tutorials.org
06.12.2024 15:07 — 👍 61 🔁 25 💬 2 📌 2
To PhD students -- if you are interested in molecular dynamics and machine learning, you can now apply for a one-month visit to our group!
Mail or DM me for more information.
fizyka.umk.pl/en/prom-eng/