We also discuss potential ways for optimizing phylodynamic analysis settings and developing more efficient algorithms for traversing the tree space. If interested, please check our preprint out! n/n
(More tree space animations of each dataset we analyzed are available at tinyurl.com/ssstreeanim.)
17.06.2025 01:25 — 👍 0 🔁 0 💬 0 📌 0
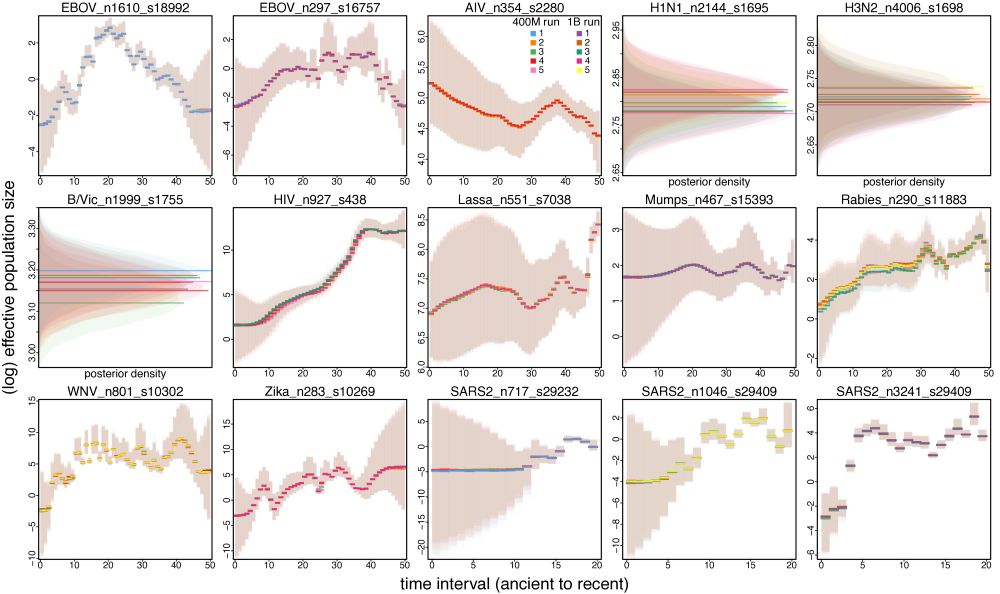
Impact of the tree sampling problems tends to be negligible on some global parameters (e.g., the effective population size through time).
Nonetheless, some global parameters governed by general tree shape are rather robust to the sampling problems. E.g., the demographic trajectories are barely distinguishable among the chains, meaning that relatively short chains would be sufficient if such parameter is of primary interest. 6/
17.06.2025 01:25 — 👍 1 🔁 0 💬 1 📌 0
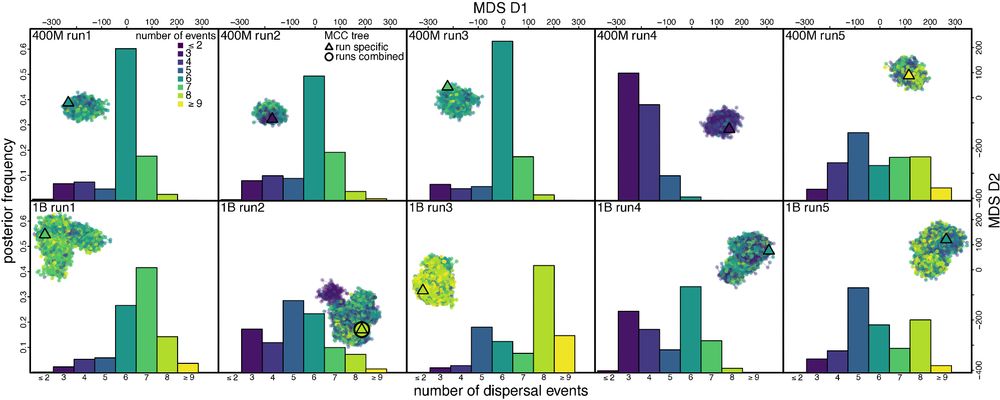
Distinct peaks in tree space can lead to different biological conclusions.
Tree space ruggedness and consequent sampling problems can strongly distort biological conclusions. E.g., our analyses of an HIV dataset reveal distinct early spread history of HIV among the chains. 5/
(The number of dispersal events between two focal geographic areas; each panel shows a chain.)
17.06.2025 01:25 — 👍 2 🔁 0 💬 1 📌 0
Our new MCMC diagnostics reveal that the sampling problems typically stem from a small part of the tree. E.g., in a LASV dataset, removing just two of the 551 sequences reshapes the tree space and effectively resolves the sampling problems. 4/
(Tree space samplers before and after the removal.)
17.06.2025 01:25 — 👍 1 🔁 0 💬 1 📌 0
We are interested in understanding phylodynamic tree landscape and its biological impacts. Here we show that the tree landscape is usually highly rugged, leading to widespread sampling problems. 3/
(MCMC progress in MDS tree space; each panel shows a dataset and each dot represents a tree.)
17.06.2025 01:25 — 👍 0 🔁 0 💬 1 📌 0
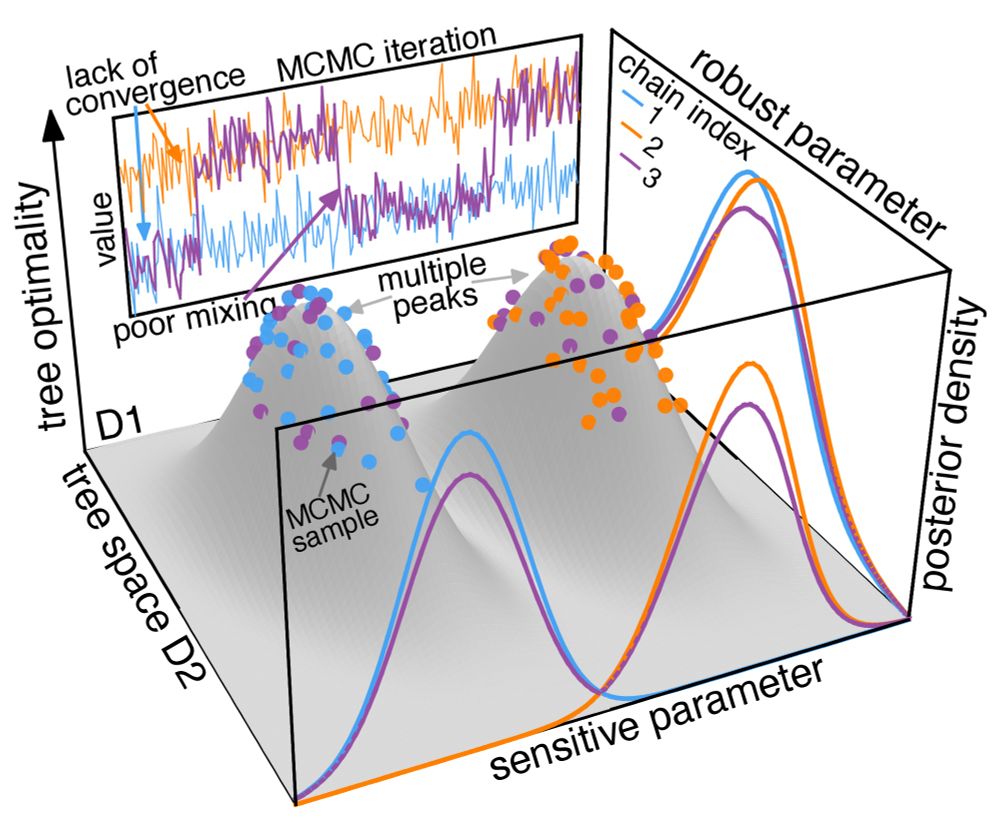
Conceptual illustration of a rugged tree landscape and the consequent sampling problems.
Unlike typical phylogenetic settings where the tree topology is our focus and the best tree is strongly supported by the informative genomic data, phylodynamic inference uses viral sequences with limited genetic diversity and frequently treats the topology as a nuisance. 2/
17.06.2025 01:25 — 👍 0 🔁 0 💬 1 📌 0
Excited to share my new preprint developed with @matsen.bsky.social, in collaboration with Marius Brusselmans, Luiz Carvalho, @msuchard.bsky.social, and @guybaele.bsky.social, on the biological causes and impacts of tree space ruggedness in phylodynamic inference. 1/
www.biorxiv.org/content/10.1...
17.06.2025 01:25 — 👍 7 🔁 8 💬 1 📌 0
An open-source pathogen sequence database dedicated to equitable sharing, transparent governance, & empowering global public health.
pathoplexus.org
Assistant Professor of Teaching in the Department of Evolution and Ecology at UC Davis.
Inclusive, evidence-based learning, quantitative reasoning in ecology education, theoretical plant ecology.
robindecker.net
Structural Phylogenetics • methods, datasets, tools, events • concise updates for researchers and practitioners. Managed by @proteinmechanic.bsky.social and @cpuentelelievre.bsky.social
Investigating the evolutionary history of genetic variations to explain biological diversity and disease in modern and ancient human populations.
sessional prof at chinese univ of hong kong (cuhk), alumni chn (fka beijing) agric univ & ubc. phylogenetics, computational molecular evolution sishuowang2022.wordpress.com
Continuous feed for phylogenetics papers from PubMed, arXiv, bioRxiv, and EcoEvoRxiv. Run by @roblanfear.bsky.social
Make your own literature bot with these instructions: https://github.com/roblanf/phypapers
UConn PhD student
Studying phylogenetics.
NSF graduate research fellow
she/her
Assistant Professor - Mississippi State University - evolutionary biologist - Penstemon
Website: benstemon.github.io
Professor & Assoc Dept Head UofA EEB (@uofa-eeb.bsky.social) in Tucson studying plant evolution, botany, polyploidy, chromosomes, and biodiversity. Views are my own. He/Him https://www.barkerlab.net
evolution & math = a super interesting job
Population and evolutionary genetics @UCDavis. Posts, grammar, & spelling are my views only. He/him. #OA popgen book https://github.com/cooplab/popgen-notes/releases
Infectious diseases & genomics. Immunologist in (voluntary) exile. Minimal sarcasm. Fierce HOA (Hater of Acronyms). Personal account - opinions expressed are my own and not those of my employer.
I’m here.
https://gkarthik.com/
Prof at #UCDavis
Work: evolution, ecology, function & phylogenomics of host-microbiome systems; #openscience;
Other: #birds; baseball; T1D
Lab phylogenomics.me
Pics jonathaneisen.smugmug.com
Links linktr.ee/jonathaneisen
TED go.ted.com/6WPm
Population geneticist, CompBio, Cornell
Full professor for Computational Evolution at ETH Zürich
Raw pu-erh in the morning; oolong in the afternoon. Chocolate: dark milk. Would rather be drawing spider genitalia, usually. Looking forward to the next field trip. Thinking about getting a pottery wheel. Father of two amazing adults. Emeritus Prof @ UBC





