
Biologically inspired graphs to explore massive genetic datasets - Nature Computational Science
A recent study proposes a data structure that addresses crucial challenges related to storage and computation of large genome databases.
📢In a recent News & Views, @ryanlayer.bsky.social discusses a data structure introduced by @aprilwei.bsky.social and colleagues for reducing storage and computational costs for phased whole-genome polymorphisms. www.nature.com/articles/s43...
🔓https://rdcu.be/d8ay3
31.01.2025 13:56 — 👍 6 🔁 2 💬 0 📌 1
Wei Lab
Web site created using create-react-app
My lab (aprilweilab.github.io) continues to develop GRG and ARG related methods & more. We are looking for a postdoc to join us.
05.12.2024 17:09 — 👍 4 🔁 5 💬 1 📌 0
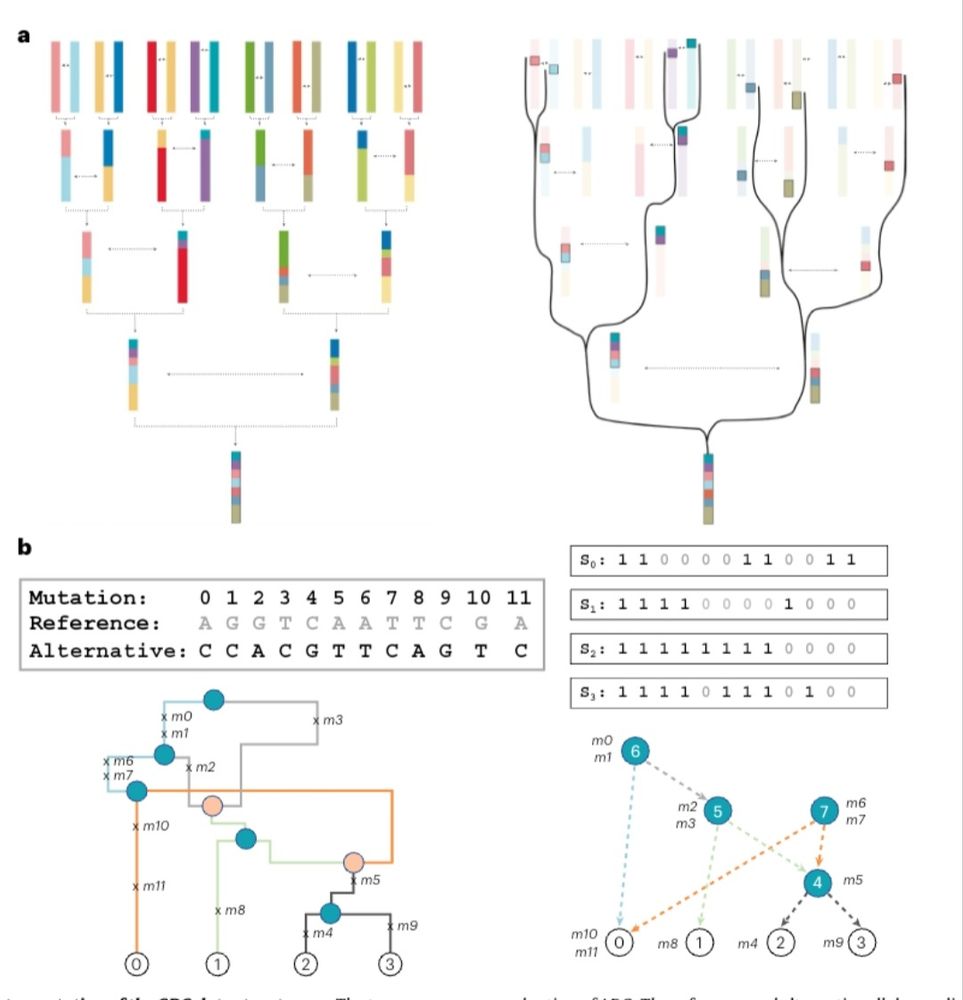
Our work w/ two co-first authors Drew DeHaas and Ziqing Pan is now published. GRG allows large amounts of WGS polymorphism data to be analyzed in RAM via graph traversal & algebra operations & has some intrinsic connection w/ popgen data generating process & is different from ARG
05.12.2024 17:09 — 👍 21 🔁 11 💬 1 📌 0
Thanks, Alison. (that's me logging in 6mo later😂
19.11.2024 22:22 — 👍 1 🔁 0 💬 0 📌 0

Genotype Representation Graphs: Enabling Efficient Analysis of Biobank-Scale Data
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
We introduced an ARG-inspired data structure, Genotype Representation Graph (GRG), to enable lossless data compression and efficient computation through graph traversal. Developed a fast inference method. Cost ~80 GBP to convert 350TB VCF (200,000 UKBiobank WGS) into 160 GB GRG.
t.co/0badfCYz47
29.04.2024 18:20 — 👍 4 🔁 0 💬 1 📌 0
PI at the Big Data Institute, University of Oxford
Royal Society University Research Fellow
aDNA | Complex Traits | Selection | Disease | Domestication
🇦🇺🇬🇧💀🧬🦠🐕
Postdoc in Pritchard lab at Stanford University. Previous PhD co-advised by Yun Song and Rasmus Nielsen at UC Berkeley.
/dee-AA-nah/|Postdoc at UCLA|computational biology PhD / popgen, genomics and conservation | she/her/ella 🇲🇽
PhD candidate in the Specht lab @Cornell
BSA student representative 2023-2025
Lover of bromeliads, genomics, reproductive biology and scicomm!
Evolutionary biologist, computational biologist, statistician. I like to develop mathematical models of evolutionary process and see how they fit to data. I also like cities where building apartments is legal.
Postdoc at Fred Hutch with @matsen.bsky.social. Phylogenetics, phylodynamics, macroevolution, molecular evolution, & Bayesian inference.
UMich professor who studies resistance and persistence (in plants)
Researcher, evolution of drug resistance, modeling, population genetics, coding, no longer at SF State University, instead at the U of Montpellier.
Mom, Dutch, now immigrant in France.
I study Neanderthals, Denisovans and the effect of their DNA surviving in present day humans. Assistant professor at section for molecular ecology and evolution at Globe Institute Copenhagen, Denmark.
Assistant Professor, University of Washington, Genome Sciences.
Previous: JSMF Fellow, Berkeley EECS
♡: Computational biology, evolutionary dynamics, quantitative immunology
https://dewitt-lab.github.io/
[disclaimer: opinions mine] 🇺🇸🚫👑
PhD student in Computational Biology at Cornell University. PopGen | EvoBio | spatial simulation | demographic history. Amateur in linguistics & calligraphy. he/his/him/هُوَ/ـهُ/他
PhD Student @ Cornell | NSF Graduate Research Fellow | popgen.dev
Evolutionary and statistical geneticist. Associate Professor at
Center for Genomic Sciences, UNAM.
http://sohaillab.com
UNAM | U Chicago | Langebio | Harvard | MIT
🇵🇰🇺🇸🇲🇽🏳️🌈
Assistant Professor at University of Michigan. Evolutionary biology, spatial population genetics, dad, he/him
Computational genomics, population genetics, data science, and AI. Former professor turned biotech researcher and consultant. Also running, gaming, and dad-ing.
https://www.alonkeinan.org/
Assistant Professor @ ASU | Studying #microbiome, #evolution, #bioinformatics, #multiomics | Developing scikit-bio (https://scikit.bio) | Open-source enthusiast
Human evolutionary genetics, University of Cambridge; Darwin College. 🇮🇪







