YouTube video by GenomeTDCC
RNAs are not naked - Gene Yeo, PhD, MBA
RNA is never naked 🧪🖥️🧬
Gene Yeo @ucsandiego.bsky.social reveals how RNA-binding proteins (RBPs) shape gene expression, drive disease, and can unlock new therapies. From splicing to synthetic RBPs, this talk is packed with insights
youtu.be/AnhbGp-pA-w
#RNA #AcademicSky
@sanfordstemcell.bsky.social
05.08.2025 17:15 — 👍 7 🔁 1 💬 0 📌 0

📢Job Opening! 🧪🖥️🧬👩🔬
Ready to hire qualified candidates to map #RNA modifications in human transcriptomes using #nanopore direct sequencing.
Dr. Ya-Ming Hou at Thomas Jefferson University
houlaboratory.com/research
#HigherEd #EDUSky #AcademicSky #STS
04.08.2025 14:28 — 👍 5 🔁 2 💬 0 📌 0
YouTube video by GenomeTDCC
The Many Lives of mRNA
Happy #RNADay!
What happens to RNA after transcription?
@stirlingchurchman.bsky.social tracks RNA flow through the cell, revealing key insights into gene regulation, nuclear degradation & RNA kinetics. A fascinating look into #RNA biology! 🧪🧬🖥️👩🔬
youtu.be/XAKMASk_fc4
01.08.2025 15:08 — 👍 6 🔁 2 💬 0 📌 0
🧪👩🔬
31.07.2025 15:13 — 👍 3 🔁 0 💬 0 📌 0
YouTube video by GenomeTDCC
A Brief History of Contemporary 3D Genomics Technologies
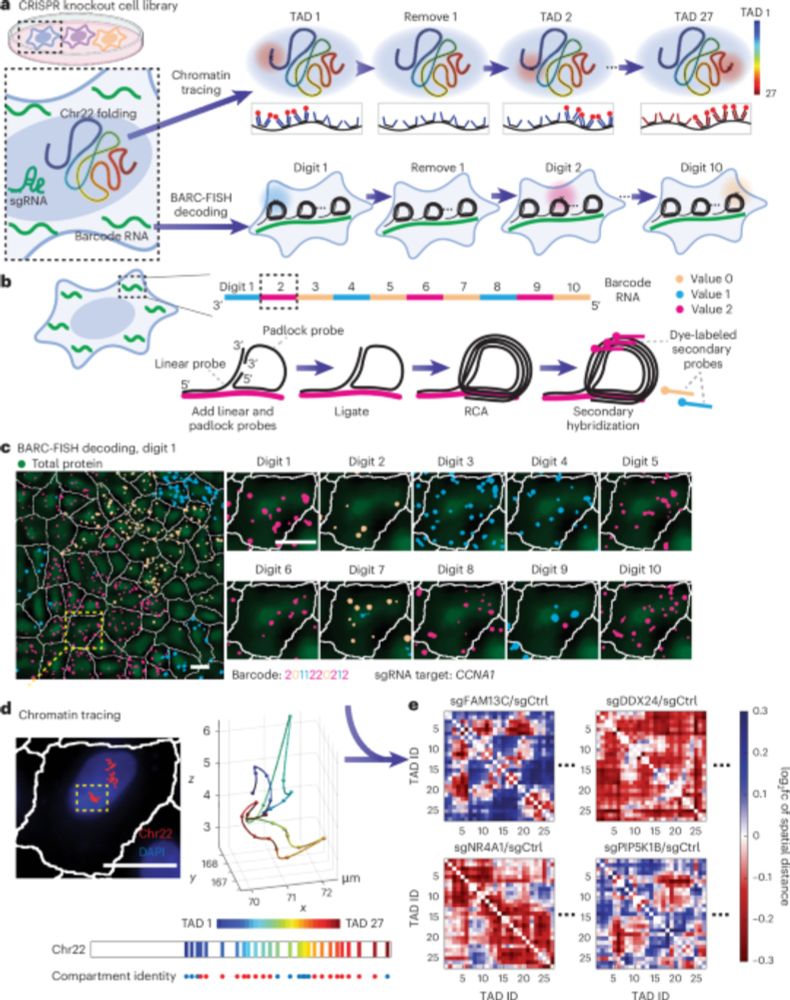
How does 6ft of #DNA fit inside a cell only 10 microns wide?
@sstevenwang.bsky.social provides a brief history of contemporary 3D #genomics technologies. Learn how spatial organization of DNA plays a vital role in gene regulation, development, and disease. 🧬🖥️🧪 #STS
www.youtube.com/watch?v=gXCO...
30.07.2025 15:53 — 👍 1 🔁 1 💬 1 📌 0

Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @cp-cell.bsky.social, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. www.sciencedirect.com/science/arti...
26.07.2025 19:15 — 👍 97 🔁 37 💬 1 📌 2

Genomic Technologies Open Science Day on June 9. The event will be held at the Eric. P. Newman Education Center at Washington University in St. Louis and broadcast live online. Registration is free at https://www.jax.org/scienceday25/
📅 June 9 - Genomic Technologies Open Science Day at @washu.bsky.social and online
www.jax.org/scienceday25
Register for Free by:
May 30: In-Person
June 8: Virtual
Learn how emerging tech is used to manipulate RNA & DNA, study genome function and structure, and navigate multi-omic analysis 🧪👩🔬🧬🖥️
28.05.2025 15:24 — 👍 2 🔁 2 💬 0 📌 0

Light blue graphic with dark blue text reads, "June 9, 2025 | Washington University, St. Louis, MO & Virtual | Genomic Technologies Open Science Day" with logos for The Jackson Laboratory, Genome TDCC and the National Human Genome Institute.
🔬 Excited about the future of genomic tech?
Join NHGRI & @genometdcc.bsky.social's hybrid forum to explore how new tools and technologies are used, as well as the potential opportunities and challenges ahead.
📅 Open to grad students, postdocs & researchers!
Register: www.jax.org/education-an...
09.05.2025 18:16 — 👍 2 🔁 1 💬 0 📌 1
HAPPY DNA DAY! 🧬🧪
Genomics research has provided humanity with a transformative understand of life at the molecular level leading to a wide range of benefits in medical advancements, synthetic biology, biotechnology, infectious disease, agriculture, and evolution.
We are connected by ATGC
25.04.2025 14:36 — 👍 4 🔁 0 💬 0 📌 0
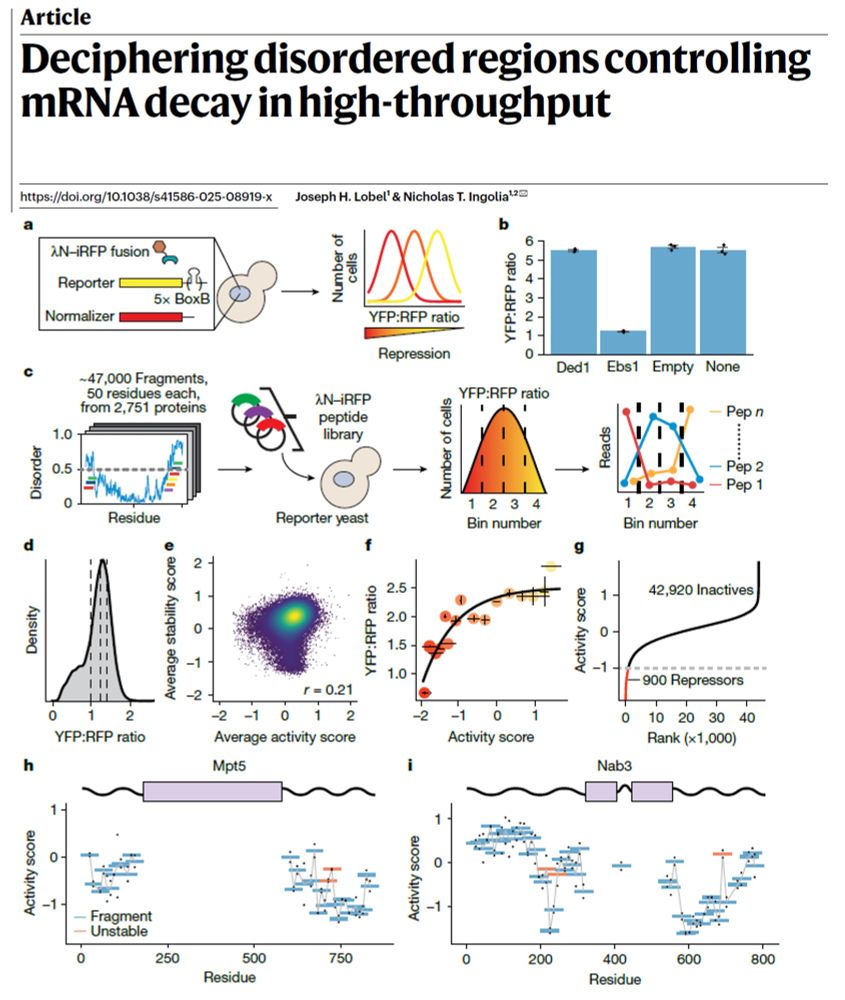
Schematic of the fluorescence-based tethering assay as described in the recently published Nature paper by Joseph Lobel and Nick Ingolia, "Deciphering disordered regions controling mRNA decay in high-throughput".
From @nickingolia.bsky.social at @ucberkeleyofficial.bsky.social 🧪🧬🖥️
Disordered protein regions hold powerful sway over gene expression, as this new large-scale study reveals how they regulate mRNA stability and translation without a fixed structure. Published in @nature.com
doi.org/10.1038/s415...
24.04.2025 12:52 — 👍 6 🔁 0 💬 0 📌 0
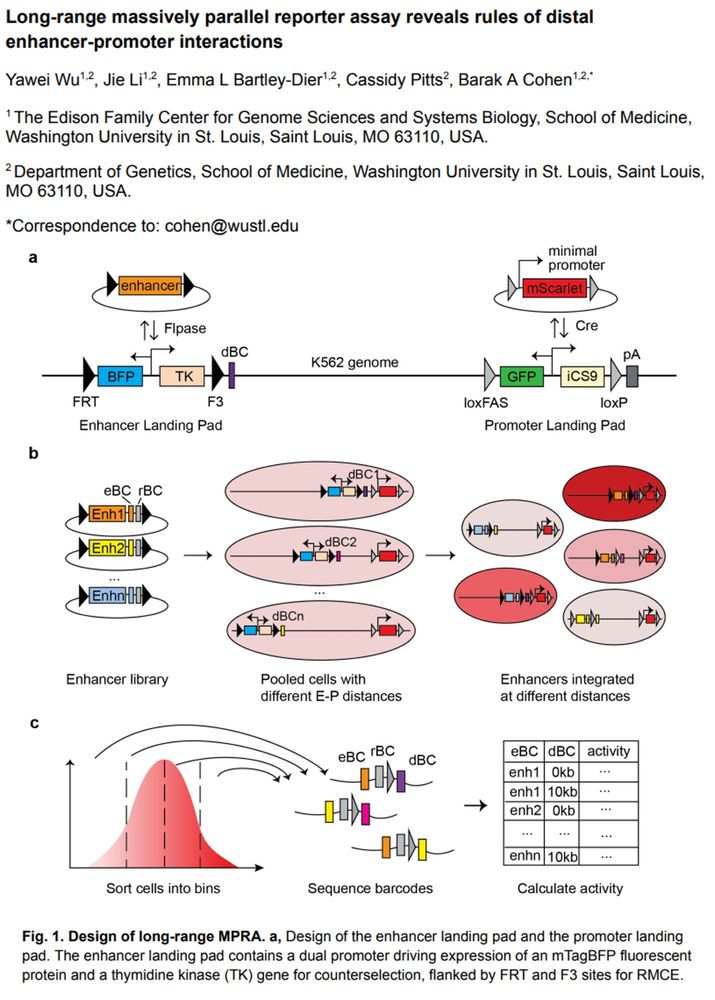
From the BioRxiv pre-print article "Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions" by Yawei Wu et al.
Fig. 1. Design of long-range MPRA. a, Design of the enhancer landing pad and the promoter landing
pad. The enhancer landing pad contains a dual promoter driving expression of an mTagBFP fluorescent
protein and a thymidine kinase (TK) gene for counterselection, flanked by FRT and F3 sites for RMCE.
available under aCC-BY-NC 4.0 International license.
was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made
bioRxiv preprint doi: https://doi.org/10.1101/2025.04.21.649048; this version posted April 22, 2025. The copyright holder for this preprint (which
The promoter landing pad expresses the mEmerald fluorescent protein and iCaspase9 for
counterselection and is flanked by loxFAS and loxP sites for orthogonal RMCE. b-c, Workflow of
long-range MPRA. eBC: enhancer barcode; rBC: random barcode; dBC: distance barcode. A barcoded
enhancer library is integrated into pooled long-range landing pad cells through Flpase-facilitated
recombination. After counterselection with Ganciclovir, cells with successful integration are enriched.
Cells are sorted into different bins based on mScarlet fluorescence level. Genomic DNA is extracted from
each bin; barcodes are amplified and sequenced. Reporter gene expression of each enhancer at each
distance is calculated as the weighted average of fluorescence levels across bins.
🚨PRE-PRINT 🧪🧬🖥️👩🔬
Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions
From Barak Cohen's lab at @washu.bsky.social
Read the pre-print 👇
doi.org/10.1101/2025...
Learn more about the research from the Cohen lab: bclab.wustl.edu
23.04.2025 20:43 — 👍 10 🔁 5 💬 1 📌 1

Free event with developers and users of genomic technologies. Register at https://www.jax.org/scienceday25/
The objective of this forum is to provide post-secondary students, postdoctoral associates/fellows and biomedical researchers with a fundamental understanding of how these new technologies are utilized, as well as emerging opportunities and challenges associated with their optimization and use.
📅 Genomic Technologies Open Science Day - June 9
FREE event featuring expert developers and users!
Register: www.jax.org/scienceday25/
Happening in person at @washu.bsky.social and online in partnership with NHGRI and @jacksonlab.bsky.social
🧪🧬🖥️👩🔬 #STS
Learn more at genometdcc.org#outreachevents
23.04.2025 15:29 — 👍 10 🔁 3 💬 0 📌 0
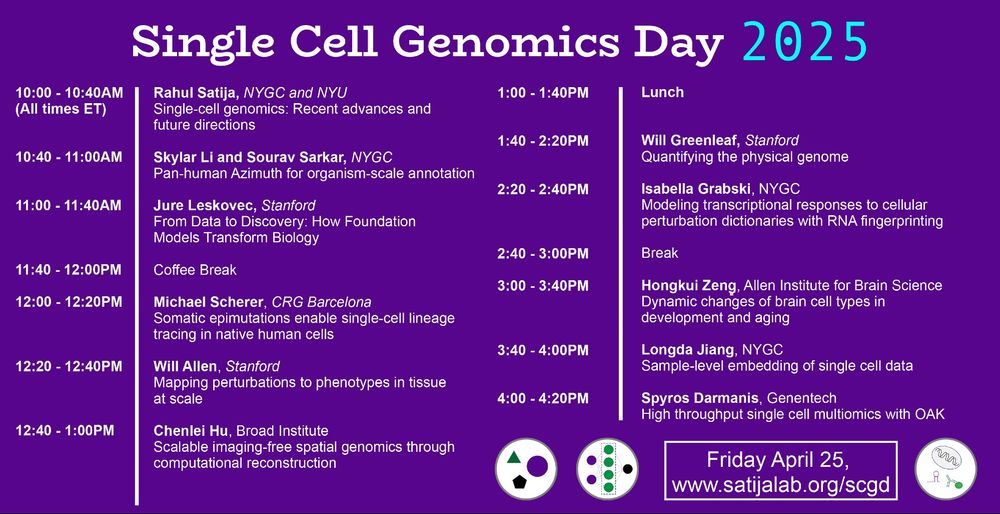
Single Cell Genomics Day hosted by Rahul Satija. Happening on April 25, 2025
Single Cell Genomics Day! 🧪🧬🖥️
Friday April 25 (10:00 AM to 5:00 PM EDT)
Watch the event live satijalab.org/scgd25/
The Rahul Satija Lab at @nygenome.org and NYU is hosting the annual SINGLE-CELL GENOMICS DAY on #DNADay
22.04.2025 13:00 — 👍 5 🔁 1 💬 0 📌 0

Graphic with dark blue and light green to light blue gradient graphic text on white background reads, "National DNA Day April 25." Alexa Wnorowski, Sarah Wojiski and Erica Gerace, JAX Genomic Education team members, are pictured standing at a table together on the left of the graphic, and Christina Vallianatos, a JAX Genomic Education team member, pipetting into a DNA helix plate on the right of the graphic.
🧬 DNA Day is April 25!
Celebrate the impact of genetics and genomics research on biomedical science with us! 🧪
Follow along this week as we highlight @jacksonlab.bsky.social’s educational programs and resources, designed to empower and inspire learners like you.
#DNADay25 #EduSky
21.04.2025 12:00 — 👍 2 🔁 1 💬 0 📌 0
Check out the 🧵 by @stanleyqilab.bsky.social on their recent publication. 🧪🧬💻 #3Dgenome
17.04.2025 19:17 — 👍 1 🔁 0 💬 0 📌 0
🧪🧬💻
17.04.2025 16:39 — 👍 7 🔁 3 💬 0 📌 0
YouTube video by GenomeTDCC
Perturb-tracing enables high-content screening of multi-scale 3D genome regulators
You can hear from the authors in this Publication Highlight video as they discuss the development of Perturb-tracing and some of their results.
youtu.be/oR63uI8feWs?...
12.04.2025 00:13 — 👍 0 🔁 0 💬 0 📌 0

The Genome TDCC YouTube channel has several videos on the latest genomic technology being developed by researchers supported by NHGRI. Highlights on new research publications and educational videos discussing new developments from nucleic acid synthesis, protein sequencing, large-scale genomic analysis and more.
DYK www.youtube.com/@genometdcc has videos on the latest genomic technology being developed by researchers supported by NHGRI? 👩🔬 🧪🧬🖥️ #STS
Highlighting new research pubs and #educational videos discussing innovations in synthetic biology, nucleic acid and protein seq, large-scale genomics, and more!
11.04.2025 19:28 — 👍 5 🔁 0 💬 0 📌 0
Research — Hou Laboratory
Postdoc Position Available 🧪
Interested in #Nanopore Direct Sequencing of #RNA?
Ya-Ming Hou at Thomas Jefferson University 👩🔬
houlaboratory.com/research
Seeking candidates with #bioinformatics skills
Send cover letter, CV, and references
For contact information, visit houlaboratory.com/contact
11.04.2025 13:57 — 👍 4 🔁 1 💬 0 📌 0
Newly published in @nature.com. Supported in part by the NHGRI Genome Technology Development Opportunity Fund. Check out the🧵 by @cfell.bsky.social below for more details about the paper.
Learn more about this and other work supported by the Opportunity Fund genometdcc.org/opportunity-...
🧪🖥️🧬 #STS
10.04.2025 20:11 — 👍 2 🔁 0 💬 0 📌 0
YouTube video by GenomeTDCC
Perturb-tracing enables high-content screening of multi-scale 3D genome regulators
Fresh off the press in @naturemethods.bsky.social
In this highlight video, Siyuan Wang and Yubao Cheng at @yale.edu unveil a new image-based high-content screening platform, Perturb-tracing, to screen for new regulators of chromatin organization at multiple length scales.
🧪🧬🖥️
youtu.be/oR63uI8feWs
10.04.2025 13:55 — 👍 0 🔁 0 💬 1 📌 0
🧪🧬🖥️👩🔬
09.04.2025 18:31 — 👍 3 🔁 0 💬 0 📌 0
📅 April 10-11: FREE VIRTUAL SYMPOSIUM
Don't miss out on this opportunity to hear from leading experts in #RNA research
View the agenda and register TODAY:
www.rnaepicenter.com/educationand...
🧪👩🔬🧬🖥️
08.04.2025 16:45 — 👍 6 🔁 2 💬 0 📌 0
🎉Congratulations to @ceesdekker.bsky.social!
Learn more about the amazing work coming from this lab at ceesdekkerlab.nl
🧪🧬🖥️ #STS #nanopore
08.04.2025 15:58 — 👍 12 🔁 2 💬 0 📌 0

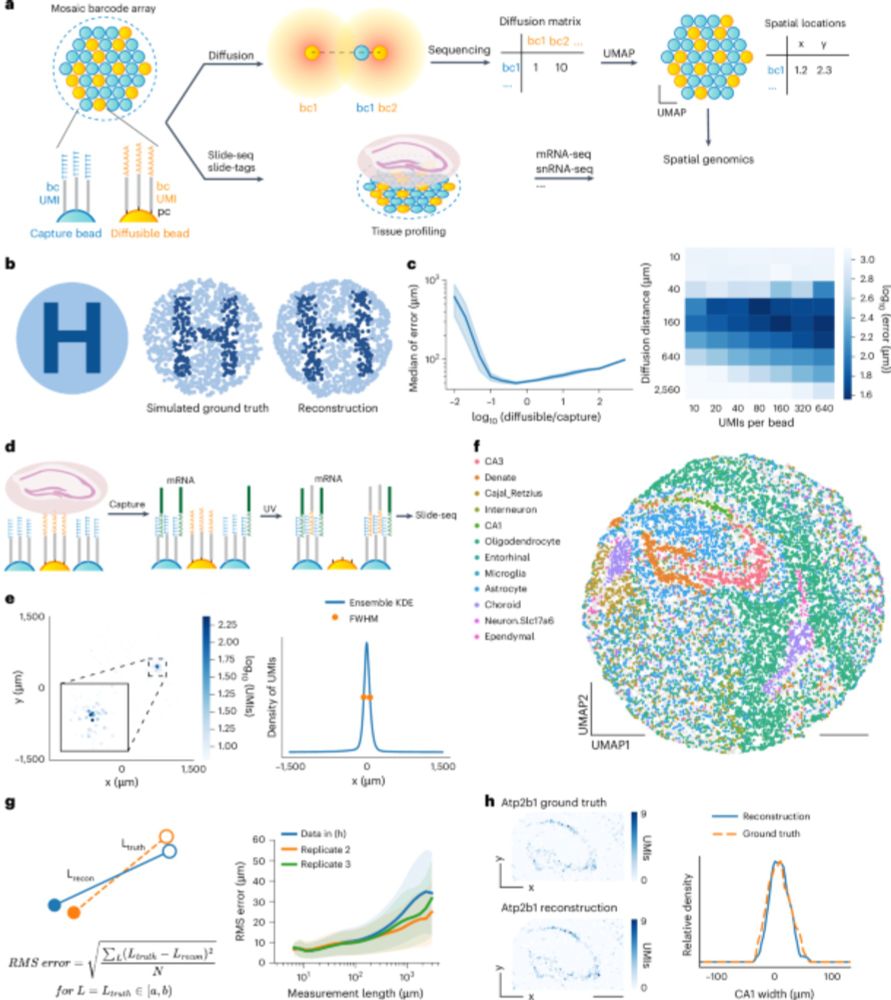
Scientists have developed a way to scale up spatial genomics and lower costs
A new computational approach eliminates time-intensive imaging, enabling high-resolution spatial mapping of gene expression in tissue.
A new spatial transcriptomics approach eliminates the need for time-intensive imaging with specialized equipment, making the technique accessible to more researchers around the world. @naturebiotech.bsky.social
03.04.2025 13:14 — 👍 28 🔁 8 💬 3 📌 0
YouTube video by GenomeTDCC
Scalable Spatial Transcriptomics Through Computational Array Reconstruction
Hear from Fei Chen and @chenleihu.bsky.social in this Publication Highlight video as they discuss this new imaging-free method.
🧪🧬🖥️👩🔬
youtu.be/dVEmk8L8ufI
04.04.2025 13:44 — 👍 2 🔁 0 💬 0 📌 0
YouTube video by GenomeTDCC
Scalable spatial transcriptomics through computational array reconstruction
Imaging-free Spatial Transcriptomics!
Computational reconstruction of spatial barcode locations using molecular diffusion + dimensionality reduction. Fei Chen and Chenlei Hu at @broadinstitute.org discuss 👇📽️
Published in @naturebiotech.bsky.social
🧪🧬🖥️👩🔬 #STS
www.youtube.com/watch?v=dVEm...
03.04.2025 12:29 — 👍 4 🔁 0 💬 1 📌 1

A two-day virtual symposium featuring global experts in RNA mass spectrometry, sharing the latest research and innovations in RNA modifications and sequencing. Hosted by the NIH/NHGRI Center for Genomic Information Encoded by RNA Nucleotide Modifications
FREE VIRTUAL SYMPOSIUM
Featuring experts in RNA mass spectrometry, sharing the latest research and innovations in RNA modifications and sequencing. April 10 - 11
Register: tinyurl.com/43kkxvp7
Hosted by the Center for Genomic Information Encoded by RNA Nucleotide Modifications
🧬🧪🖥️ #STS #RNA
28.03.2025 13:08 — 👍 8 🔁 1 💬 0 📌 1
The UC San Diego Sanford Stem Cell Institute is a global hub for stem cell science and innovation, advancing research in space & through active clinical trials.
Sanford Labs is an independent, not-for-profit biomedical research organization with a mission to discover and deliver the next generation of molecular medicines
The official journal of the RNA Society. Featuring cutting edge research in RNA biology.
https://rnajournal.cshlp.org
🧬 The Genome • Unrestricted 🧪
Focused on innate immune mechanisms governing pathogenesis of disease and on NK-cell based therapies for disease. Opinions expressed are Dr. Waggoner's alone and not on behalf of lab staff or employer.
Join us in uniting the Genomics community! Our goal is to deliver the benefits of genomics to patients faster.
The goal of the Canadian BioGenome Project is to produce high-quality reference genomes 🧬 for all Canadian species 🌎
Sequencing Canada's Biodiversity 🌿🦋🐍🧬🐢🦌🐸🌳🐿🐙🦈🦦
Learn more: https://linktr.ee/canadianbiogenome
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
An international online community about chromatin and gene regulation.
Fragile Nucleosome community: Discord forum, seminars and more
Discord: http://discord.gg/dXqT89r
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Guest Scientist IMP Vienna, Board of Directors NumFOCUS
Incoming Prof UMass Chan Medical
Previously Stanford Genetics, UW CSE.
Asst. Prof. at WashU School of Medicine; biophysics/biochem/evolution of intrinsically disordered proteins. How does nature encode function without a stable structure? We work in vivo / in vitro / in silico. He/him.
https://www.holehouselab.com/
Professor at the NYU School of Medicine (https://yanailab.org/). Founder and Director of the Night Science Institute (https://night-science.org/). Co-host of the 'Night Science Podcast' https://podcasts.apple.com/us/podcast/night-science/id1563415749
Assistant professor @ Duke University BME, guerilla knitter, pipettor of small volumes of liquid. Biologist masquerading as engineer, or the other way around.
Penn Epigenetics Institute at The University of Pennsylvania Perelman School of Medicine, Philadelphia, PA
Asst Prof at University of California, Irvine.
Genetics, Genomics, Gene Regulation, Development. Views are my own.
https://www.kvonlab.org/
Studying genome organization with quantitative super-resolution microscopy | Postdoc at Harvard/Wyss | PhD at MPIB and LMU
https://www.researchgate.net/profile/Johannes-Stein-4
Postdoc in Anders Hansen lab @ MIT | PhD from Barak Cohen lab @ WUSTL | Gene regulation ❤️ | 🇸🇬
Department of Genetics at Washington University School of Medicine
Hiring faculty&postdocs: https://genetics.wustl.edu/positions/
Postdoc in the Shendure & Pinglay labs @UW
Trying to understand and engineer our wonderfully weird genomes 🧬