Seminars | Activities | KIAS(Korea Institute For Advanced Study)
85 Hoegi-ro, Dongdaemun-gu, Seoul 02455, Republic of Korea.
If you happen to be in Seoul tomorrow Dec. 3 don't miss this seminar at the Korea Institute for Advanced Study (KIAS)
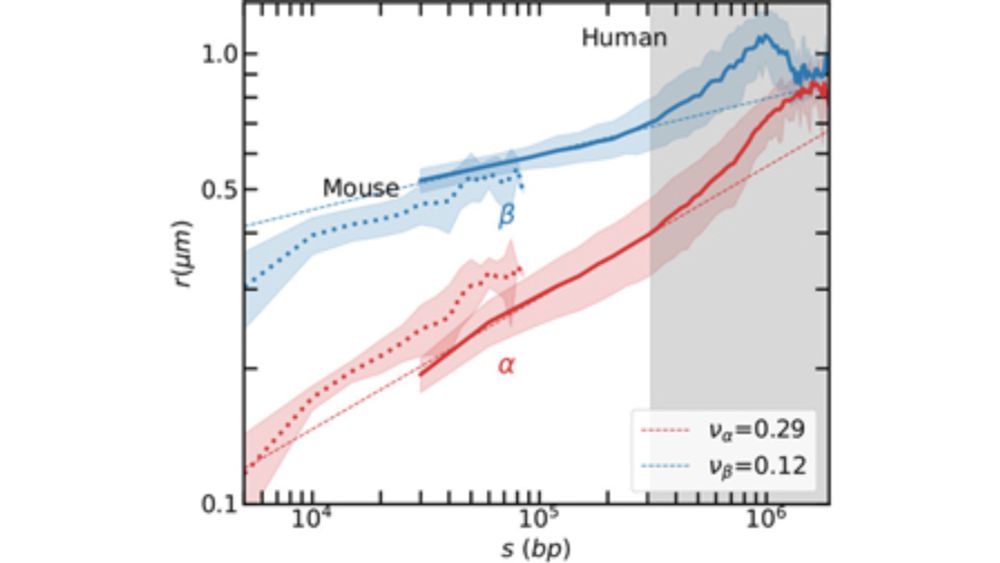
"Chromatin structure from high resolution microscopy: Scaling laws and microphase separation" by Midas Segers (KU Leuven)
www.kias.re.kr/kias/activit...
02.12.2025 20:07 — 👍 0 🔁 0 💬 0 📌 0
RNAScale
Exploring RNA Dynamics across Scales
🎉Our group has been awarded a prestigious #FIS grant! #RNAScale - Exploring RNA Dynamics across Scales: From Base Pairs to Viral Genomes 👉 www.bussilab.org/fis3
We will soon open postdoctoral positions across several components of the project! An expression-of-interest form is already online.
28.11.2025 13:35 — 👍 16 🔁 7 💬 1 📌 2
In this project, we will investigate chromosome architecture by integrating theoretical modelling, numerical simulations, and the analysis of experimental data.
An exciting journey ahead — I look forward to our collaboration.
#statistical_physics #polymers #chromatin #MD_simulations
04.11.2025 15:01 — 👍 3 🔁 0 💬 0 📌 0

Congratulations to Giandomenico Vanore on being awarded a PhD Fellowship by @fwovlaanderen.bsky.social
He will conduct his doctoral research under my supervision on the project “Understanding the structure of interphase chromosomes.”
#statistical_physics #polymers #chromatin #MD_simulations
04.11.2025 15:01 — 👍 4 🔁 0 💬 1 📌 0

Working on biosensors? Don’t miss this MDPI webinar introducing thermal biosensing via the heat-transfer method (HTM) — a simple yet powerful way to detect biomolecular interactions and monitor cells.
Pioneers of HTM will share their insights!
#Biosensors #HTM
More info: sciforum.net/event/Sensor...
08.10.2025 12:16 — 👍 1 🔁 1 💬 0 📌 0

🧬 Seminar Alert!
Join us next Friday 17 Oct. for “Mechanics, Dynamics and Shape in Oocytes” by Dr. Bart Vos — a deep dive into how physical forces and cell mechanics shape early development.
Discover the emerging field of mechanobiology, where physics meets life.
14.10.2025 16:20 — 👍 1 🔁 0 💬 0 📌 0

Interest in pursuing a PhD/postdoctoral position in experimental Condensed Matter Physics with applications to biological systems? Don’t miss this opportunity from the "Small Biosystems Lab" felixritortlab.com UBarcelona which combines statistical physics with single-molecule experimental techniques
14.10.2025 08:58 — 👍 2 🔁 0 💬 0 📌 0

Compounding formula approach to chromatin and active polymer dynamics
Active polymers are ubiquitous in nature, and often kicked by persistent noises that break detailed balance. In order to capture the out-of-equilibrium dynamics of such active polymers, we propose a s...
New preprint! With Takahiro Sakaue we studied an active polymer dynamics focusing on the mean-square displacement of a monomer. We show that the MSD can be described by a simple compounding formula linking an isolated monomer MSD with tension propagation along the chain -
arxiv.org/abs/2510.07949
12.10.2025 16:03 — 👍 0 🔁 0 💬 0 📌 0

MeChaNiSM
Mechanical Characterization of Nucleic acids using Single Molecule techniques
Traveling back after an exciting three days meeting of the MeChaNiSM Doctoral Network @mechanism-network.bsky.social - Thanks to all students, PIs and supporting staff for these intense days of scientific discussions about our projects on DNA/RNA mechanics. www.chalmers.se/en/projects/...
10.10.2025 12:10 — 👍 1 🔁 0 💬 0 📌 0

Inferring DNA kinkability from biased MD simulations
In several biological processes, such as looping, supercoiling and DNA-protein interactions, DNA is subject to very strong deformations. While coarse-grained models often approximate DNA as a smoothly...
New preprint out! We investigate DNA kinks using biased all-atom MD simulations, in collaboration with Arianna Fassino and Aderik Voorspoels @aderikvoorspoels.bsky.social, as part of the EU-funded Doctoral Network MeChaNiSM @mechanism-network.bsky.social
www.biorxiv.org/content/10.1...
14.09.2025 06:51 — 👍 1 🔁 1 💬 0 📌 0

📢Seminar 11 September at 12h, Faculty of Science, Seminar Room Module 5
@cienciasuam.bsky.social @uam.es
Enrico Carlon @ecarlon.bsky.social from KU Leuven
will talk about DNA mechanics beyond the twistable wormlike chain. See you there!
ifimac.uam.es/ifimac-semin...
04.09.2025 08:12 — 👍 9 🔁 4 💬 0 📌 1

Applicants wanted 5:
Open position for PhD Student Studying Vitro Single-molecule Transport Across Peroxisome Mimics
careers.tudelft.nl/job/Delft-Ph...
22.08.2025 08:52 — 👍 3 🔁 2 💬 0 📌 0

Palazzo Franchetti. Istituto Veneto di Scienze, Lettere ed Arti
Enjoying this workshop on the Statistical Mechanics of polymers in Venice these days...
Great program and organization!
userswww.pd.infn.it/~orlandin/Ve...
21.07.2025 12:19 — 👍 2 🔁 0 💬 0 📌 0
Collaboration with Midas Segers @pol-kuleuven.bsky.social, Enrico Skoruppa and Helmut Schiessel @poldresden.bsky.social (4/4)
22.04.2025 18:43 — 👍 0 🔁 0 💬 0 📌 0
In these experiments a single DNA molecule is attached to a solid surface and to a bead which is pulled and rotated by magnetic fields. Our paper develops a theory of multiplectonemes formation in stretched and twisted DNA, which is in excellent agreement with simulations. (3/4)
22.04.2025 18:43 — 👍 0 🔁 0 💬 1 📌 0

Plectonemes are supercoiled structures that form when DNA is overwound (or underwound). They help DNA fit in tight space and play an important role in gene regulation. The figure illustrates plectonemes forming in DNA under torque and tension as in magnetic tweezers in vitro experiments. (2/4)
22.04.2025 18:43 — 👍 0 🔁 0 💬 1 📌 0
Very happy that our latest publication "Statistical mechanics of multiplectoneme phases in DNA" doi.org/10.1103/Phys... was selected as Editors' Suggestion. These suggestions list "a small number of papers that the editors and referees find of particular interest, importance, or clarity". (1/4)
22.04.2025 18:43 — 👍 6 🔁 1 💬 2 📌 0

Exciting news! PoL@KUL invites you to the inaugural lecture of the International Francqui Professor 2024-25, Prof. Davide Marenduzzo (U. of Edinburgh) on Tuesday, April 29, 2025 in Leuven:
Biophysical principles of chromosome organisation
Info and registration:
fys.kuleuven.be/english/coll...
03.04.2025 13:50 — 👍 9 🔁 3 💬 0 📌 1

MeChaNiSM
Mechanical Characterization of Nucleic acids using Single Molecule techniques
Welcome to Arianna Fassino who has recently joined our group to start a PhD project on simulations of Nucleic Acids Mechanics. The project is part of a EU-Marie Skłodowska Curie Doctoral Network “MeChaNiSM” Mechanical Characterization of Nucleic acids - www.chalmers.se/en/projects/...
31.03.2025 09:14 — 👍 4 🔁 0 💬 0 📌 0

Apply now for our international Master of Science in Physics of Life @biozentrum.unibas.ch @unibasel.ch! Scholarships available.
More information: www.biozentrum.unibas.ch/education/de...
#Master #Students #Research #Physics #Mathematics #ComputerScience #Engineering #Basel #Switzerland
20.03.2025 08:10 — 👍 33 🔁 19 💬 0 📌 4
Very pleased to share our new story on SMC complexes, a great team effort from @roisnehamelinf.bsky.social, @taschner.bsky.social in the GruberLab and James of @dnatopologylab.bsky.social We find that SMC hinge domains are ssDNA gates to facilitate obstacle bypass during DNA loop extrusion!
18.03.2025 08:03 — 👍 15 🔁 5 💬 2 📌 0

schematic model for obstacle navigation by SMC rings by extrusion through the SMC lumen and by hinge bypassing
Now, 20+ years later, our lab revisits the hinge using the anti-plasmid SMC complex Wadjet. We show that the SMC hinge is a DNA gate—an obstacle bypass gate selective for ssDNA, used to navigate chromosomal barriers during DNA loop extrusion.
18.03.2025 07:56 — 👍 2 🔁 1 💬 2 📌 0
Yet another preprint from the lab—this one is personal. During my PhD, I ended up working on the cohesin SMC hinge, showing its role in sister chromatid cohesion and proposing it as a DNA entry gate in the SMC ring. But the details have remained mysterious and controversial.
18.03.2025 07:55 — 👍 44 🔁 10 💬 3 📌 3
Hinge bypass can explain some otherwise-difficult to explain observations made by colleagues using single molecule imaging and by simulating chromosome folding. DNA loop-extruding SMC complexes turn out to be even a bit more wonderful.
18.03.2025 07:57 — 👍 2 🔁 1 💬 0 📌 0
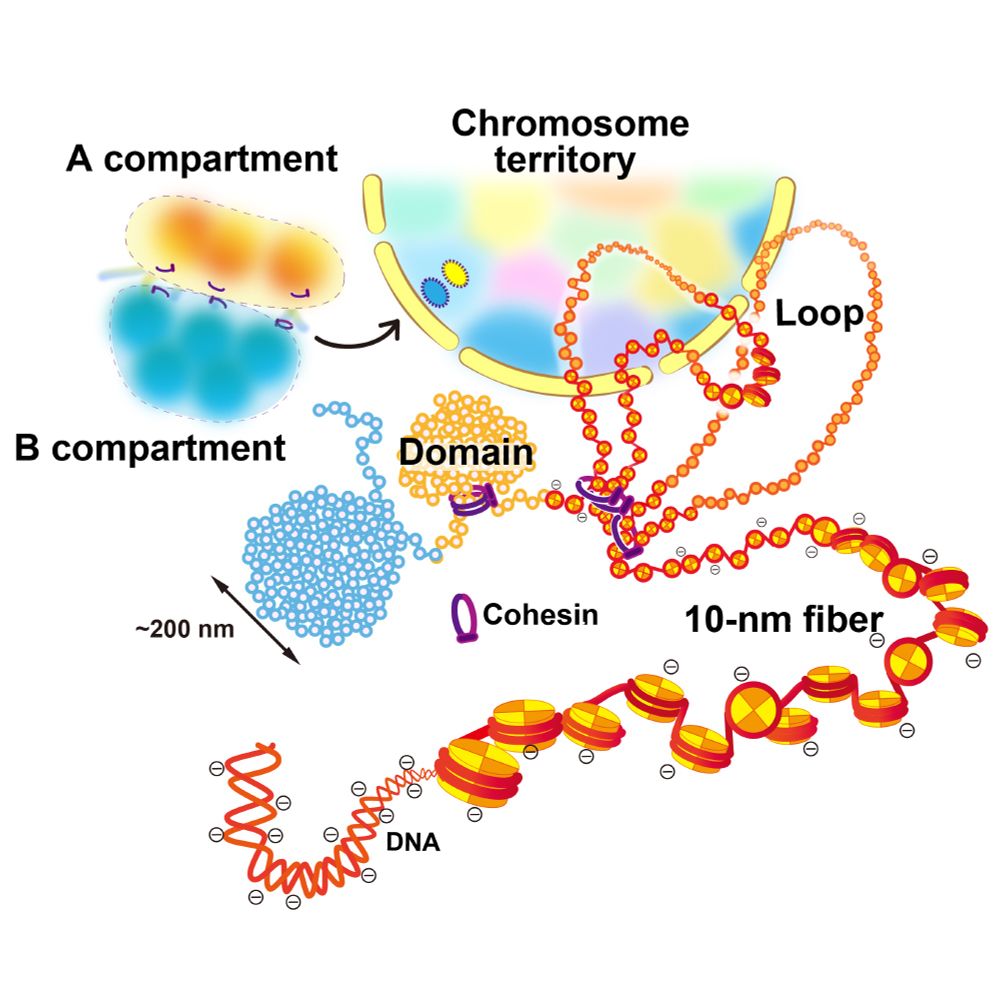
Our new review on how the #chromatin domain is formed in the cell is now available @Curr Opin Struct Biol.📄✨
We critically discuss the domain formation mechanism from a physical perspective, including #phase-separation and #condensation.
📥 Free-download link:
authors.elsevier.com/a/1keGn,LqAr...
20.02.2025 11:14 — 👍 75 🔁 37 💬 0 📌 0
Research Fellow at the Flatiron Institute. Incoming Assistant Professor of Physics at University of Toronto 2026 | Previously: IBM Research, Cornell Physics | Statistical mechanics, theoretical biophysics, dynamical systems
davidhathcock.github.io
Research, Education and Business. Vienna BioCenter is a life science cluster that includes IMP, IMBA, GMI, Max Perutz Labs, as well as the Faculty of Life Science & CeMESS (both University of Vienna) and 35+ biotechs.
PhD student at Ghent University.
Interested in complexity, information theory and networks.
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, AI/ML in biotech // http://albertvilella.substack.com
Vice Rector International Policy, Interculturality and Alumni Policy - Professor in Experimental Physics - KU Leuven
Prof (CS @Stanford), Co-Director @StanfordHAI, Cofounder/CEO @theworldlabs, CoFounder @ai4allorg #AI #computervision #robotics #AI-healthcare
Meet new people
Researcher working w innovation. Interests: #Climate (esp health & climate) #LifeScience #Politics #Security #Sustainability. NOT INTERESTED IN CRYPTO OR OF! Born @310 ppm Due to current situation in the world I consider myself a dystopian optimist.
Mathematics Sorceror (sensory alchemist) at the Arctangent Transpetroglyphics Algra Laboratory (ATAL), I transflarnx mathematics into living rainbows. http://owen.maresh.info https://github.com/graveolensa
Psoeppe-Tlaxtlal, (an undreamt splendour?)
Marie Curie PhD candidate @UvA | IIT Kharagpur '25 | Computational Chemistry Enthusiast
Professor at University of Cologne, Germany.
Organic Chemistry, Chemical Biology, Nucleic Acids
Biophysicist. Lab website: http://artemefremovlab.com/
EU-funded network connecting 8 academic laboratories with 4 industrial partners to train 15 doctoral students.
We are the Ting Wu Lab @ HMS. Our laboratory studies how chromosome behavior and positioning influence genome function, with implications for gene regulation, genome stability, and disease.
Managed by WuLab members
https://www.transvection.org/home
Computational Physicist @ University of Cambridge | physical chemistry | polymers | soft matter
WEB: https://sites.google.com/view/romanstano/
Fluorescence, RNA, DNA, RNA therapeutics, spectroscopy, microscopy, optical tweezers, nucleic acid conformation and dynamics
Interested in #physics of life. Theorist but some of my best friends are experimentalists.
Research Assistant ll @ Georgia Tech, School of Biological Sciences, @McGrath's Lab
Coming "PhD student @Fox Lab, CWRU
Biophysicist tying to make sense of life.
Interdisciplinary @FWOVlaanderen PhD Fellow with @ColindaScheele at @vib_ccb.