End-to-end protein design in the browser through evedesign. Generate and interactively explore designs in 2D/3D and export them as codon-optimized DNA. The underlying open source framework (released soon) is build to easily add new methods, more on that soon.
🌐 evedesign.bio
22.10.2025 14:30 — 👍 91 🔁 30 💬 2 📌 1

Looking for a new approach to studying or eliminating phages? Check out our study introducing anti-phage ASOs (antisense oligos) out in @Nature today. nature.com/articles/s4158…
10.09.2025 15:40 — 👍 133 🔁 65 💬 4 📌 2
hey bluesky 👋 visa hurdles mean I’m looking for opportunities outside the US. I’m a computational biologist (bacterial + phage genomics, postdoc in Koonin’s group @ NIH). I am interested in teaming up on funding apps. reach out if this resonates!
15.09.2025 17:26 — 👍 70 🔁 91 💬 1 📌 3

RNAcanvas: interactive drawing and exploration of nucleic acid structures
Abstract. Two-dimensional drawing of nucleic acid structures, particularly RNA structures, is fundamental to the communication of nucleic acids research. H
🧪 #RNAsky
If you aren’t using RNAcanvas to draw your RNA structures and explore alternative structures, you are missing out! 56 citations in a year, with multiple ones in top journals like Nature, Science & Cell. Easy to use and packed with unique features! Try it!
academic.oup.com/nar/article/...
28.08.2025 22:33 — 👍 63 🔁 17 💬 1 📌 1

Happy to share our paper in Nano Letters!🧬 We developed programmable RNA nanostructures for detecting cotranscriptionally introduced RNA modifications using nanopores. Big congrats to first authors Iva &
Gerardo and thanks to all collaborators! 🔗https://pubs.acs.org/doi/10.1021/acs.nanolett.5c02391
04.08.2025 17:41 — 👍 3 🔁 3 💬 1 📌 0
Employing "a much larger ribozyme" is always the solution to any problem, in my opinion.
03.07.2025 19:49 — 👍 0 🔁 0 💬 0 📌 0
Thank you so much for sharing! I'm glad you like it!
03.07.2025 10:58 — 👍 1 🔁 0 💬 1 📌 0
RNAcanvas
A web app for drawing and exploring nucleic acid structures.
Thank you! Yes, absolutely! We wouldn't know what to do without it. I strongly recommend people check it out! www.rnacanvas.app
02.07.2025 18:58 — 👍 2 🔁 2 💬 1 📌 0
Yes, that's correct, and honestly I was surprised with how well it worked, too. I believe the trick was paying close attention to sequence/structure motif conservation which seems to hold sufficient information to reconstruct the whole ribozyme class.
02.07.2025 15:26 — 👍 1 🔁 0 💬 0 📌 0
I'm grateful to my student Lukas, who did a lot of the early characterization of Arq.I2, and of course @mutschlerlab.bsky.social for his mentorship and making this project possible! And ofc @crc392molevo.bsky.social!
I will be answering any questions, so feel free to ask if you have any! (11/11)
02.07.2025 10:01 — 👍 1 🔁 0 💬 1 📌 0
This project was my brainchild and I'm overjoyed to finally see it published!
I want to thank my co-1st author: @noemi-n.bsky.social for all her hard work in seeing the project through until the end! I expect a lot of cool upcoming work from her, so follow her if you like group II introns! (10/11)
02.07.2025 10:01 — 👍 2 🔁 0 💬 1 📌 0
This work serves as a proof of concept for the viability of complex ncRNA design through inverse folding, and we hope it paves the way for the computational design of bespoke G2Is and possibly other complex ncRNAs with traits that are favorable for biotech or therapeutic applications. (9/11)
02.07.2025 10:01 — 👍 0 🔁 0 💬 1 📌 0

Surprisingly, we saw modest GFP expression compared to a control with an inactive intron. G2Is tend to silence host genes by cleaving the mRNA, so lowered expression is not unexpected. These results indicate that the protein-free intron is active in E. coli cells. (8/n)
02.07.2025 10:01 — 👍 0 🔁 0 💬 1 📌 0

According to conventional wisdom, G2Is require protein cofactors to aid in self-splicing under intracellular conditions, hindering their application as protein-free genome editors.
We designed an assay for splicing-dependent fluorescent protein expression — a GFP gene interrupted by Arq.I2 (7/n)
02.07.2025 10:01 — 👍 1 🔁 0 💬 1 📌 0

Unlike typical de novo designed ribozymes, Arq.I2 outperforms most wild type variants characterized thus far! It reacts with fast kinetics, having a rate of splicing comparable to those of the fastest known natural G2Is! (6/n)
02.07.2025 10:01 — 👍 0 🔁 0 💬 1 📌 0

All three candidates were active in vitro, some even outperforming a variant of a natural intron.
Arq.I2 is a very unusual intron — it requires extreme measures to denature it for PAGE, and remains catalytically active in the absence of monovalent cations. (5/n)
02.07.2025 10:01 — 👍 0 🔁 0 💬 1 📌 0

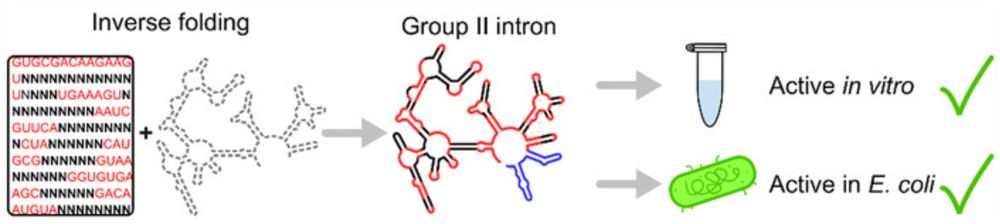
In our latest paper we combined the inverse folding algorithm aRNAque and rational design to generate unique synthetic G2Is.
aRNAque's evolutionary algorithm was able to finetune intron folding, resulting in unusually stable structures. (4/n)
02.07.2025 10:01 — 👍 1 🔁 0 💬 1 📌 0
Protein-free G2Is were recently found to hydrolyze dsDNA, showing potential as RNA-only genome editors (see my previous thread). However, their complexity poses a challenge to their de novo design, as only short ribozymes have ever been generated using inverse folding algorithms. (3/n)
02.07.2025 10:01 — 👍 0 🔁 0 💬 1 📌 0
G2Is are self-splicing retroelements found in all domains of life, and are the ancestors of the spliceosome.
Owing to dozens of tertiary interactions and a large conformational change between the steps of catalysis, they are among the largest and most complex ribozymes. (2/n)
02.07.2025 10:01 — 👍 0 🔁 0 💬 1 📌 0
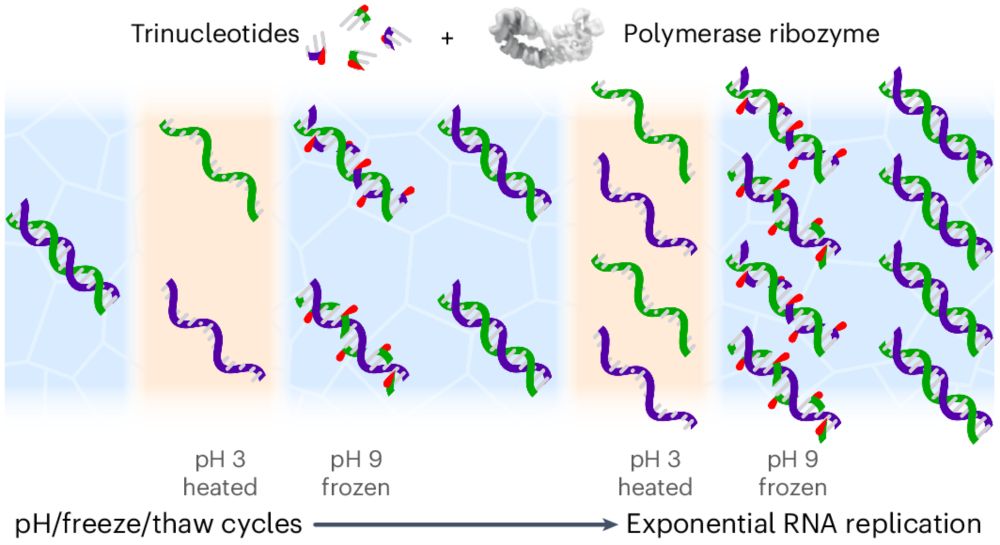
Computational De Novo Design of Group II Introns Yields Highly Active Ribozymes
Group II Introns (G2Is) are large self-splicing ribozymes with promising biotechnological applications. This study utilized RNA inverse folding to design three novel G2Is. The designed intron Arq.I2,...
Have you ever wanted to design large and complex ribozymes from scratch? I know I have!
We used RNA inverse folding to design synthetic Group II Introns (G2Is) from the bottom up, resulting in highly active #ribozymes that splice in E. coli without proteins!
#intron #RNAsky #SynBio 🧬🧵(1/n)
02.07.2025 10:01 — 👍 23 🔁 9 💬 3 📌 2

A general substitution matrix for structural phylogenetics.
Abstract. Sequence-based maximum likelihood (ML) phylogenetics is a widely used method for inferring evolutionary relationships, which has illuminated the
New paper from the lab from Sriram Garg in my group. We introduce a general substitution matrix for structural phylogenetics. I think this is a big deal, so read on below if you think deep history is important. academic.oup.com/mbe/advance-...
11.06.2025 14:01 — 👍 96 🔁 52 💬 3 📌 2

An intron endonuclease facilitates interference competition between coinfecting viruses
Introns containing homing endonucleases are widespread in nature and have long been assumed to be selfish elements that provide no benefit to the host organism. These genetic elements are common in vi...
The preprint mentions that you find thousands of hits in UniProt for these new HEGs. Do you know if these are standalone HEGs, or if they are predominantly associated with introns?
Could these HEGs block coinfection of competing phages, similar to what was found for group I introns?
26.05.2025 15:24 — 👍 1 🔁 0 💬 0 📌 0
Christmas came early this year! I have been wondering about this for years!
26.05.2025 11:43 — 👍 0 🔁 0 💬 0 📌 0
Emancipated Archaea 🦠
Nowadays, H₂-dependent archaea rely on partnerships with organisms that produce hydrogen. But on early Earth, when life began, archaea were probably on their own. So, where did they get the H₂ from? Let’s dig into the science.
#Science #OriginOfLife
(1/5)
20.05.2025 09:31 — 👍 18 🔁 7 💬 1 📌 0
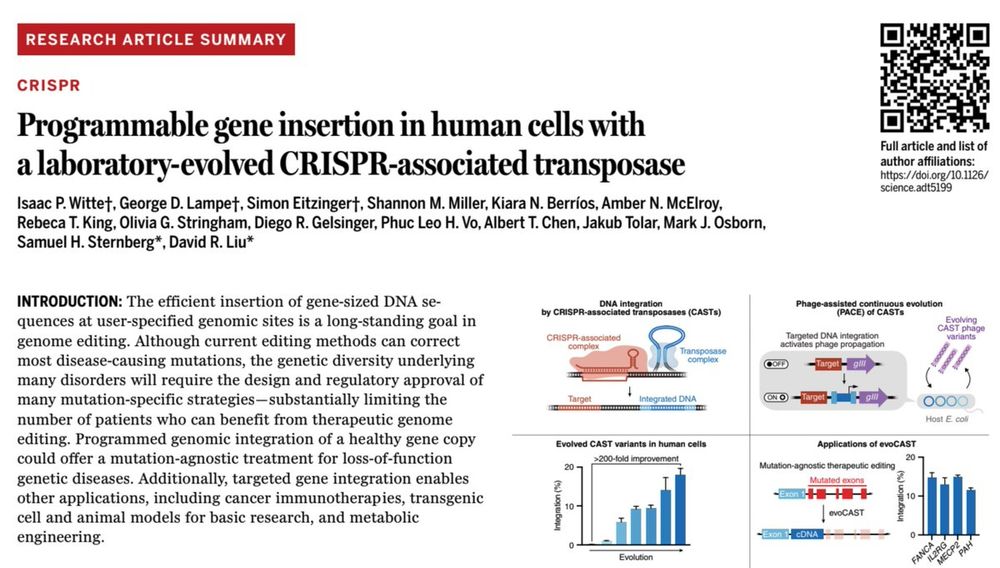
1/10 Today in @science.org in collaboration with
the Liu group we report the development of a laboratory-evolved CRISPR-associated transposase (evoCAST) that supports therapeutically relevant levels of RNA-programmable gene insertion in human cells. drive.google.com/file/d/1I-Ub...
15.05.2025 22:49 — 👍 132 🔁 59 💬 2 📌 5
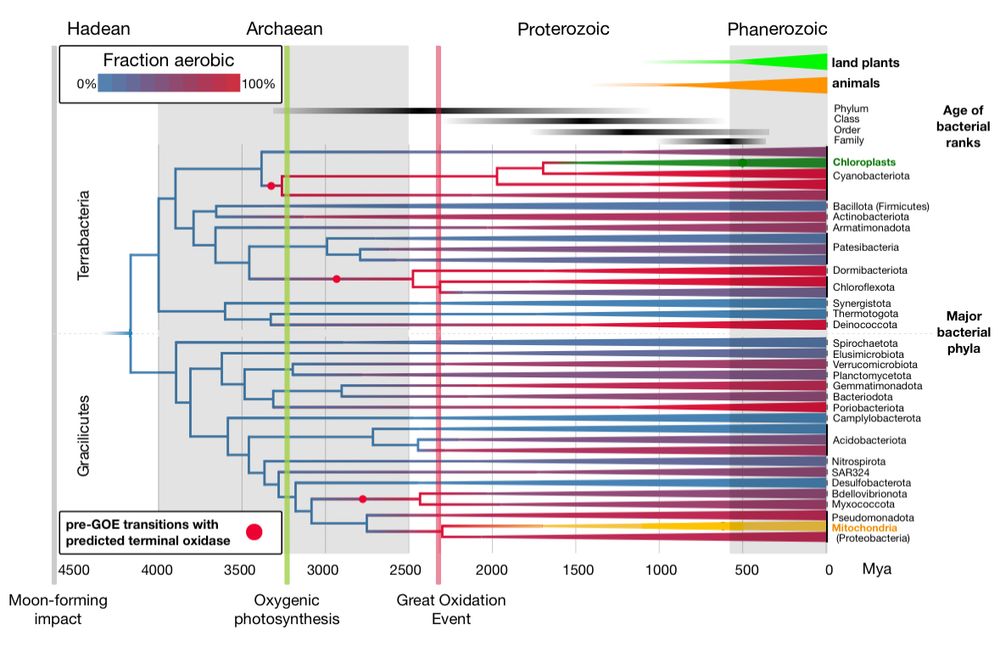
Dating Bacteria is hard due to the lack of maxima. Assuming aerobes likely postdated the GOE gave us better resolved ages, but also surprised us, but not Dr Dayhoff, showing O2 use predated its atmospheric rise by 900 Mys and helped oxygenic photosynthesis to evolve. www.science.org/doi/10.1126/...
04.04.2025 01:20 — 👍 43 🔁 26 💬 8 📌 4
Biomolecular designer
Assistant Professor at the Institute of Science and Technology Austria (ISTA)
Seminar on Protein Structure Evolution every 2nd Tue, 4 PM GMT.
Link: https://tinyurl.com/prose-seminar2
By @claudiaalcar.bsky.social @caro-rocha.bsky.social @lacholt.bsky.social @zachary-ardern.bsky.social @romeritos.bsky.social @hassanzaman.bsky.social
Mobile Genetic Elements | CRISPR-Cas | Phage-Host interactions
@csic.es researcher on the frontiers of life, freediver and RNA lover
Biophysics PhD Student with DasLab & ChiuLab.
Pairing RNA structure & cryoEM to better understand both!
King's College, Science & Security '19
Harvey Mudd '18
RNA, microbiology, infectious disease; Helmholtz Institute for RNA-based Infection Research & University of Würzburg, Germany
www.helmholtz-hiri.de
Inventing the future of biology with chemistry and biophysics. EMBO Long-Term Fellow at the Szostak Lab, HHMI & University of Chicago. Former Winton, Schiff, and Benefactors’ Scholar at the University of Cambridge’s Keyser Lab. 🧬🧪🕳️🔬
Group Leader at Helmholtz Munich and TUM (MIBE) | Formerly PostDoc/Visiting Scholar in Adam Cohen's (@adamezracohen.bsky.social) lab at Harvard | Synbio, Gene Reporters, Integrators and Recorders for High-Res Imaging and Molecular Actuators
Assistant Prof @UBChemistry | Computational modeling of RNA structure and folding, intrinsically disordered proteins and phase separation | BioSimLabUB.github.io
scientist at UC Berkeley inventing advanced genomic technologies
lover of molecules, user of computers
https://scholar.google.com/citations?user=63ZRebIAAAAJ&hl=en
Network of excellent European research institutions that have joined forces to collaborate on synthetic cell research and accelerate its translation into technology
Professor University of Maryland, co-founder Silvec Biologics finding RNA solutions to plant diseases, love Plant Virology, author Real Science Behind The X-Files, and passionate about dressage!
PostDoc at EEP-Lille. Interested in the repetitive, but complex, fraction that make up most of the genomes. Fan del Sol
Excited to engineer a synthetic cell from scratch with lipid vesicles and DNA/RNA origami-based molecular hardware. Professor @ZMBHh @UniHeidelberg
Postdoc working on RNA Nanotech & MD simulations @uniheidelberg.bsky.social & MPIP. GROMACS wrangler, oxDNA developer, @molpigs.bsky.social podcast host, and all around weird lil guy.
Has been known to post about music and neat bugs found in the woods.
PhD Student @ https://niopeklab.de/
Engineering switchable proteins💡
Views and opinions are my own
🇺🇸 in 🇩🇪
CNRS researcher at IRCAN (Nice, France) Université Cote d'Azur (UniCA) - Interested in retrotransposons, mobile DNA, transposable elements and jumping genes
About TEs, IFN, senescence with a zest of ML
🇫🇷 Parisian gravitating in the Pacific Northwest 🇺🇲
-> Seattlelite🗼🛸👾