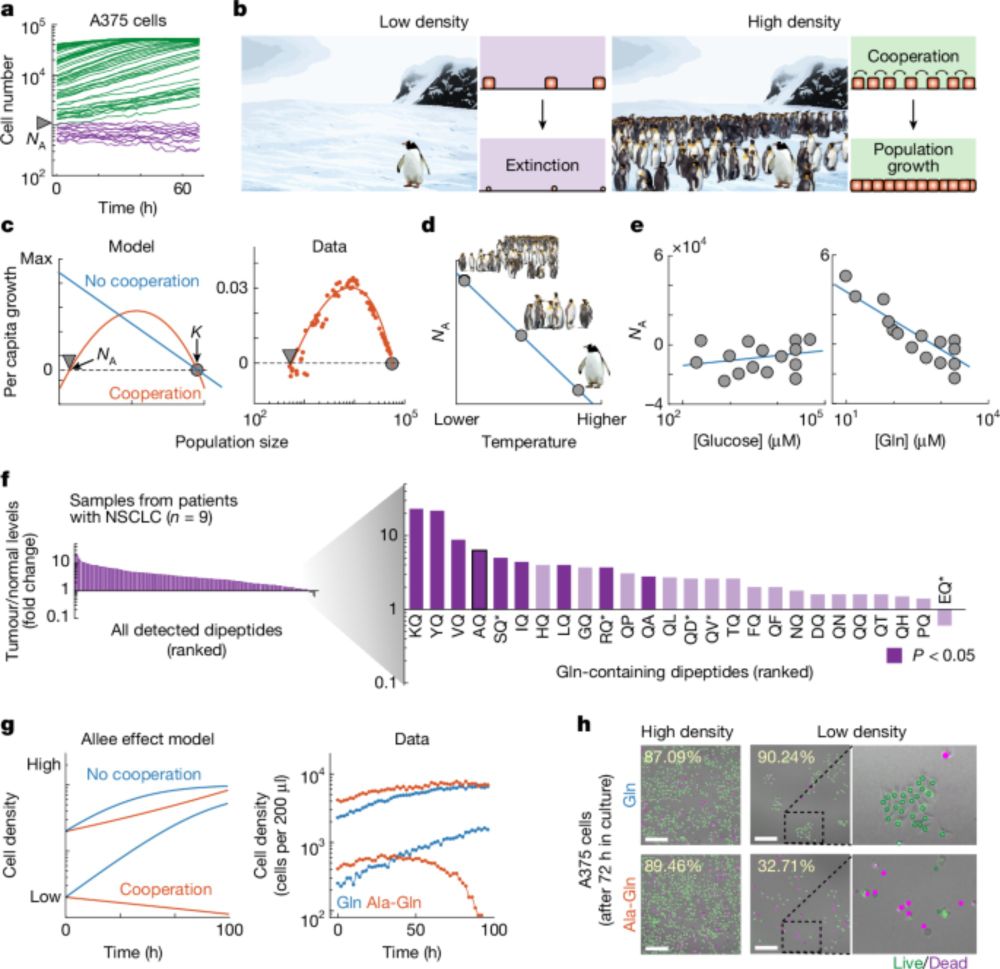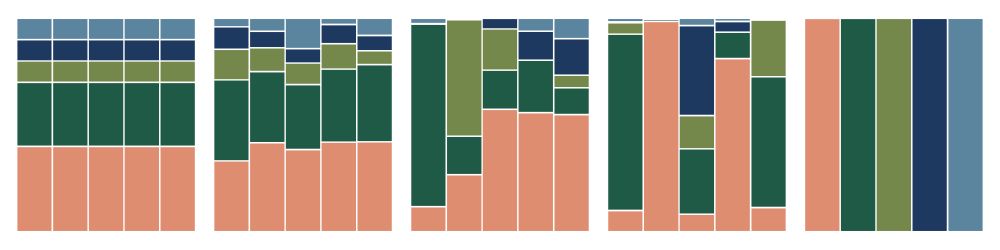
5 relative abundance plots arranged to have increasing compositional variability (variability across relative abundance samples, here vertical bars)
1/ Hey y'all, I'm excited to share my latest paper, which is out now in PNAS! We introduce FAVA, a statistical framework to measure compositional variability across microbiome samples. If you want to measure variability across a stacked bar plot, FAVA is for you! Paper: doi.org/10.1073/pnas...
14.03.2025 20:46 — 👍 207 🔁 73 💬 5 📌 2

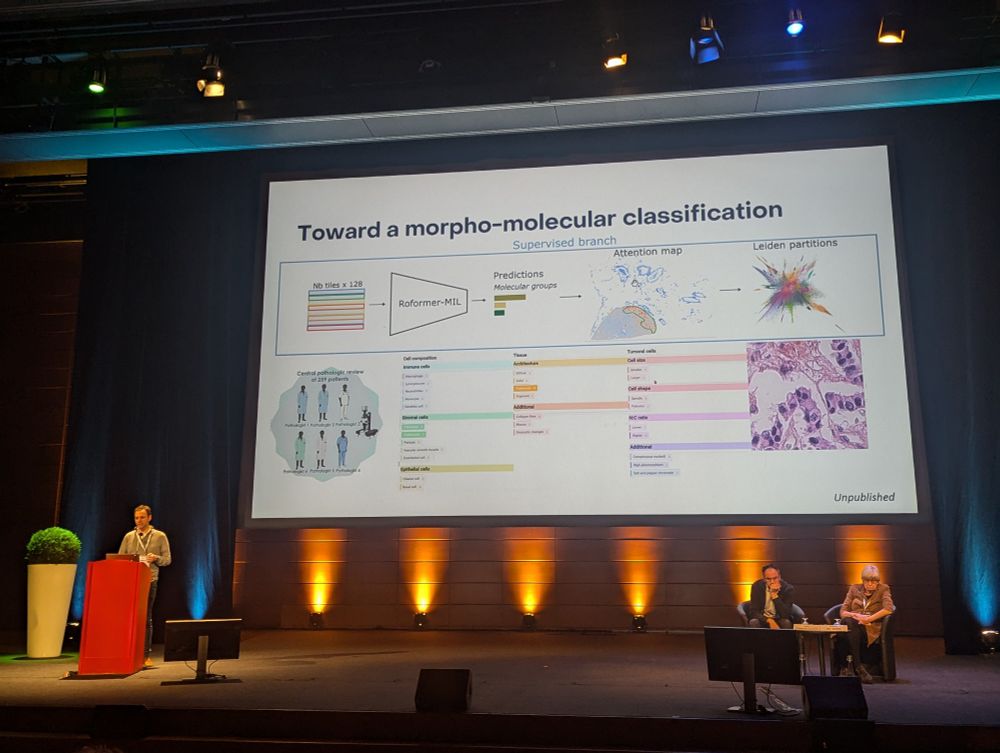
Inspiring talk from @matthieufoll.bsky.social and Lynnette Fernandez-Cuesta about the future of #lung #neuroendocrine #tumor classification and clinical management, and how to use #AI to learn simple morphological features to recognize molecular subtypes #CRCL25
31.01.2025 16:07 — 👍 4 🔁 2 💬 0 📌 0

@lisemangiante.bsky.social from @stanfordpress.bsky.social : breast #cancer adopt specific ecological strategies to adapt to their microenvironment 🤯 #CRCL25
29.01.2025 15:00 — 👍 3 🔁 1 💬 0 📌 0
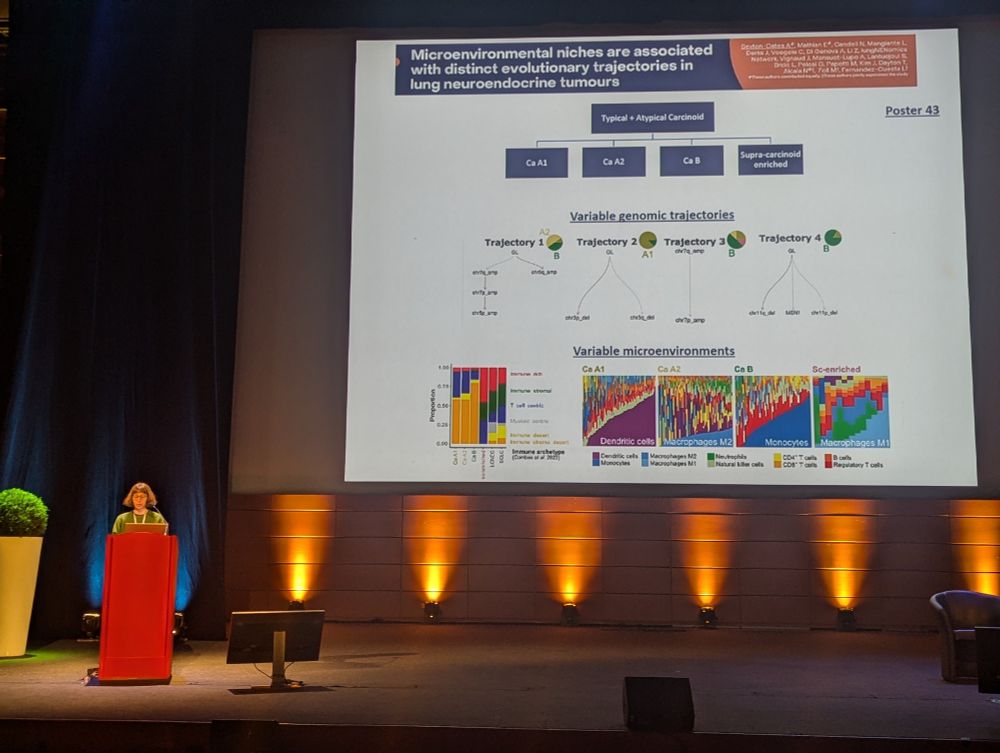
How do lung #neuroendocrine #tumors and their microenvironment coevolve ? Great talk by Alexandra Sexton Oates at #CRCL25 #LungNETs #lungcancer
29.01.2025 11:33 — 👍 4 🔁 2 💬 0 📌 0
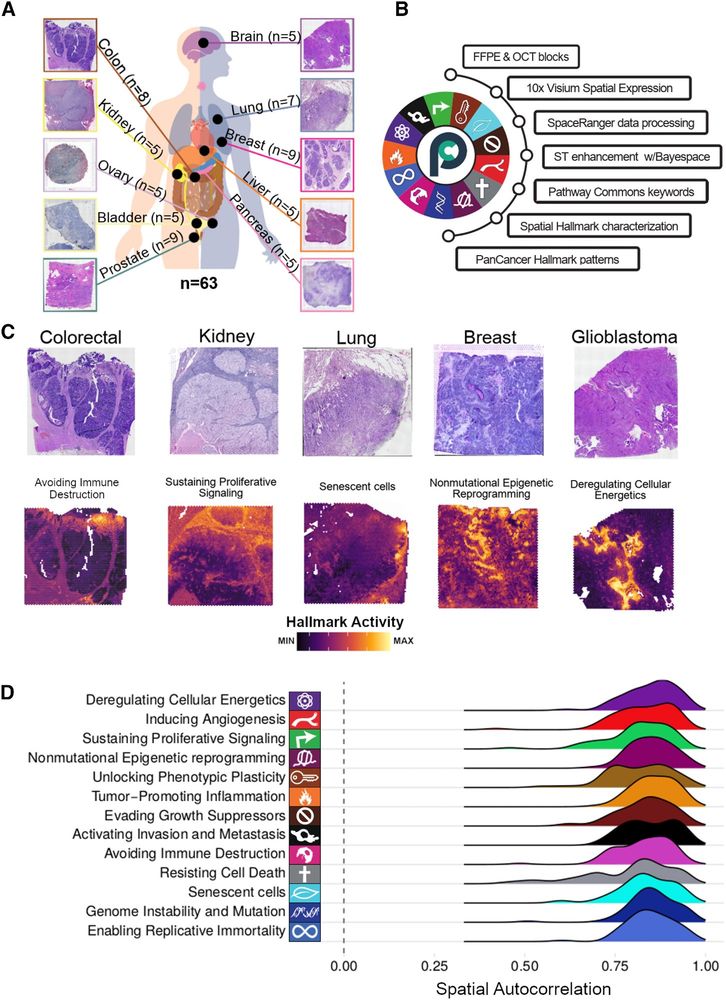
Thrilled to introduce our Cell Reports @cellreports.bsky.social article, showing how the iconic 'Hallmarks of Cancer' are located in the space of real human primary tumors. A team effort co-led by Eduard Porta, Matthew H. Bailey, and yours truly #SpatialBiology #Visium
www.cell.com/cell-reports...
25.01.2025 17:07 — 👍 8 🔁 5 💬 1 📌 0
🚨 Very proud of this commentary led by @rijazaidi.bsky.social reasoning on the impact of methods and experimental design on cancer evolution understanding, motivated by the outstanding work of @TheBoutros, @VanLooLab, @kellrott et al
Original work 👉 doi.org/10.1038/s415...
Our Commentary 👇
16.12.2024 15:47 — 👍 1 🔁 1 💬 1 📌 0
Recommendations for Bioinformatics in Clinical Practice www.biorxiv.org/content/10.... "Key recommendations include adopting the hg38 genome build as the reference and a standard set of recommended analyses, including the use of multiple tools for structural variant (SV) calling"
06.12.2024 14:15 — 👍 2 🔁 1 💬 0 📌 0
Congratulations @EMathian! So well deserved! https://x.com/CancersRare/status/1738201813545848991
22.12.2023 14:19 — 👍 0 🔁 0 💬 0 📌 0
Vacancies
Still looking for a postdoc? Yes, we have a second position to join our team and work with @nl_alcala @FCLynnette and myself! https://rarecancersgenomics.com/vacancies/
#PostdocJobs https://x.com/CancersRare/status/1726698337007776217
20.11.2023 20:33 — 👍 0 🔁 0 💬 0 📌 0
Vacancies
Looking for a postdoc? Join our team to work with @nl_alcala @FCLynnette and myself! https://rarecancersgenomics.com/vacancies/
#PostdocJobs https://x.com/CancersRare/status/1726697028372348929
20.11.2023 20:31 — 👍 0 🔁 0 💬 0 📌 0
https://x.com/cancersrare/status/1636469351212634112
16.03.2023 20:50 — 👍 0 🔁 0 💬 0 📌 0
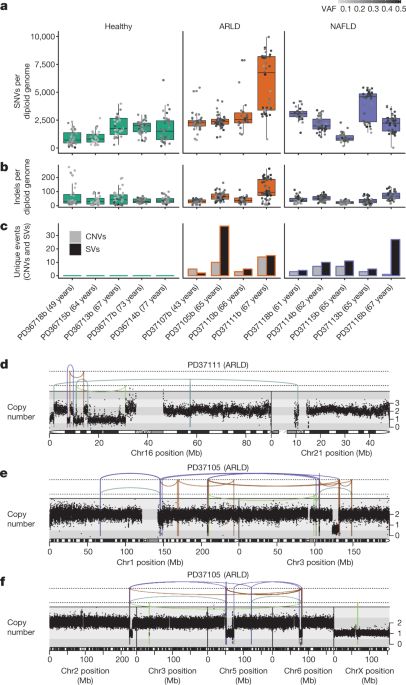
So happy to see this paper published and feeling lucky to lead the #MESOMICS project together with @FCLynnette and a fantastic team @CancersRare. Cancers associated with asbestos exposure are deadly and rising, despite its classification as a carcinogen by @IARCWHO 50 years ago https://t.co/FId6...
16.03.2023 17:21 — 👍 0 🔁 0 💬 1 📌 0

Proton and alpha radiation-induced mutational profiles in human cells
Ionizing radiation (IR) is known to be DNA damaging and mutagenic, however less is known about which mutational footprints result from exposures of human cells to different types of IR. We were interested in the mutagenic effects of particle radiation exposures on genomes of various human cell types, in order to gauge the genotoxic risks of space travel, and of certain types of tumor radiotherapy. To this end, we exposed cultured cell lines from the blood, breast and lung to intermittent proton and alpha particle (helium nuclei) beams at doses sufficient to affect cell survival. Whole-genome sequencing revealed that mutation rates were not overall markedly increased upon proton and alpha exposures. However, there were changes in mutation spectra and distributions, such as the increases in clustered mutations and of certain types of indels and structural variants. The spectrum of mutagenic effects of particle beams may often be cell-type and/or genetic background specific. Overall, the mutational effects of recurrent exposures to proton and alpha radiation on human cells appear subtle, however further work is warranted to understand effects of chronic, long-term exposures on various human tissues.
### Competing Interest Statement
The authors have declared no competing interest.
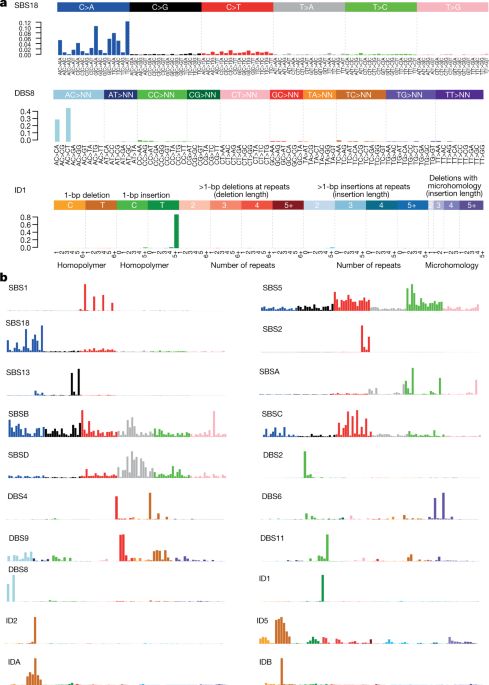
Hesitating between being proud or jealous that former PhD student @tm_delhomme’s latest preprint very first sentence mentions “extraterrestrial colonization”… Sorry I missed it during summer! I https://www.biorxiv.org/content/10.1101/2022.07.29.501997v1
12.10.2022 19:08 — 👍 0 🔁 0 💬 0 📌 0

Disentangling heterogeneity of Malignant Pleural Mesothelioma through deep integrative omics analyses
Malignant Pleural Mesothelioma (MPM) is an aggressive cancer with rising incidence and challenging clinical management. Using the largest series of whole-genome sequencing data integrated with transcriptomic and epigenomic data using multi-omic factor analysis, we demonstrate that MPM heterogeneity arises from four sources of variation: tumor cell morphology, ploidy, adaptive immune response, and CpG island methylator phenotype. Previous genomic studies focused on describing only the tumor cell morphology factor, although we robustly find the three other sources in all publicly available cohorts. We prove how these sources of variation explain the biological functions performed by the cancer cells, and how genomic events shape MPM molecular profiles. We show how these new sources of variation help understand the heterogeneity of the clinical behavior of MPM and drug responses measured in cell lines. These findings unearth the interplay between MPM functional biology and its genomic history, and ultimately, inform classification, prognostication and treatment.
![Figure][1]</img>
### Competing Interest Statement
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organisation, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organisation. Where authors are identified as personnel of the Centre de Recherche en Cancerologie de Lyon (CRCL), the authors declare no conflict of interests. A.S. participated in expert boards and clinical trials with Astra-Zeneca, BMS, MSD, Roche. N.G. declares consultancy, research support from BMS, Astra-Zeneca, Roche, and MSD. All the other authors declare no conflict of interests.
[1]: pending:yes
So proud of this preprint https://biorxiv.org/cgi/content/short/2021.09.27.461908v1, read this thread to know more: https://x.com/CancersRare/status/1443249588261949440
29.09.2021 16:24 — 👍 0 🔁 0 💬 0 📌 0
Great work from the @CancersRare team: @gabriel_aurelie @LiseMangiante @nl_alcala and @FCLynnette
04.11.2020 20:17 — 👍 0 🔁 0 💬 0 📌 0
Open science = open access + open data + open code + open peer review. Thanks @GigaScience for allowing all this and the great publication experience. Cc @usenextjournal https://x.com/GigaScience/status/1324079575878557697
04.11.2020 20:10 — 👍 0 🔁 0 💬 1 📌 0
Playing with @github #Codespaces beta. First thing tried is of course running @nextflowio!
02.10.2020 17:25 — 👍 0 🔁 0 💬 0 📌 0

nextflow hello
Running @nextflowio in @usenextjournal in a snap of a finger: https://nextjournal.com/mfoll/nextflow-hello
15.10.2019 12:58 — 👍 0 🔁 0 💬 0 📌 0
Great place, great project, great mentoring, speaking from experience! https://t.co/o8iY8MFuCN
30.09.2019 07:46 — 👍 0 🔁 0 💬 0 📌 0
The three technologies bioinformaticians need to be using right now http://www.opiniomics.org/the-three-technologies-bioinformaticians-need-to-be-using-right-now/
13.08.2019 08:07 — 👍 0 🔁 0 💬 0 📌 0
We are organising a @nextflowio training in September at @IARCWHO in #Lyon together with @canceroCLARA, please RT. #HPC #Bioinformatics https://t.co/cQa9gUdiyv cc @LyonDataScience @grenobledata @JeBiF @IFB_Bioinfo @SfbiFr @BioinfoFr
09.07.2019 08:05 — 👍 0 🔁 0 💬 0 📌 0
Executive Editor/Team Leader Open Access Science Journals Sage Publishing
Opinions = mine
http://linkedin.com/in/jlovick-editor
#oncology #cancerresearch #medicine #biology #cardiology #neurology #microbiology #publichealth #healthcare #technology
CNRS Researcher - France - Europe
Evolutionary Biology - Genomics - Ecology
Group leader and Royal Society URF @Wellcome Sanger Institute
Branco Weiss fellow @University of Cambridge
speciation, genomics, hybridisation
Computational biologist, associate professor at University of Geneva, Department of Pathology and Immunology. Single-cell omics, bio data science, immunology & cancer research
Professor in Systems Biology & Genetics @EPFL, opinions my own; Single Cell Omics / Gene Regulation / Transcription Factor / Stem Cells / Regulatory Variation / ML / Imaging / Adipose Biology / Microfluidics
https://www.epfl.ch/labs/deplanckelab
Prof of Computer Science and Life Sciences @ EPFL Previously @ Stanford
ML🤖+Bio🧬 https://brbiclab.epfl.ch/
The SIB Swiss Institute of Bioinformatics is an internationally recognized non-profit organization dedicated to biological and biomedical #DataScience.
We provide essential ressources, expertise and services and represent Swiss bioinformatics.
PI @ Cancer Research Centre of Lyon.
Bioinformatician, interested in the dynamics of tumour evolution and heterogeneity
Vice-président Métropole de Lyon
1er vice-président SYTRALMobilités
Président SAGYRC
#RareDiseases #multipleEndocrineNeoplasiaType1 #EuropeanMENAlliance www.emena.eu
#CovidIsNotOver #ImpfenRettetLeben
#MaskeAuf
Cancer epigenetics, CNRS/Cancer Research Center of Lyon (CRCL)
I do research in evolutionary genomics at @cpgsthlm.bsky.social and @stockholmuni.bsky.social #ancientDNA #mammoths #popgen #pollinators #conservation #genomics #museomics
👉 www.palaeogenetics.com/people/david-diez-molino/
don’t tell me you aren’t
✉️ you can also dm me your fav sci memes to post here
🔬 Stanford Postdoc
🏥 Stanford Medicine - 💊 Anesthesiology
🎓 Nextflow Ambassador | Sigma Xi Member
🧬 Bioinformatician
📍 California, USA
🌐 https://www.atomodyssey.com
An open-access journal from society-owned publisher, Bioscientifica. Publishing research and reviews on the interplay between hormones and cancer, and related topics. Visit eo.bioscientifica.com to learn more.
Exploring #ComputationalBiology & #Bioinformatics + Documenting my learning journey | Built by @noureldenrihan.bsky.social | 🌐 djosergenomics.github.io
Account of the Cellosaurus (www.cellosaurus.org) knowledge resource, the encyclopedia of cell lines
Watch our introductory video on: https://www.youtube.com/watch?v=xKA2AleIe0g
Cancer researcher at Hunter College of the City University of New York & Weill Cornell Medicine. Views are my own.
Your daily dose of hope, inspiration, and information in the fight against cancer.
oncodaily.com