#BPS2026 The T&C subgroup is hosting a fun event on Saturday evening, Feb 21st 🎉 Open bar for 2 hours + food! Tickets are selling fast, so sign up soon: www.biophysics.org/2026meeting/...
10.02.2026 17:18 — 👍 2 🔁 3 💬 1 📌 1#BPS2026 The T&C subgroup is hosting a fun event on Saturday evening, Feb 21st 🎉 Open bar for 2 hours + food! Tickets are selling fast, so sign up soon: www.biophysics.org/2026meeting/...
10.02.2026 17:18 — 👍 2 🔁 3 💬 1 📌 1
🌟 New Tool Alert!
We present MemPrO – a toolkit for orienting and building membrane protein systems:
✅ Membrane protein orientation
✅ Mixed membrane building
✅ Double membranes & curvature
✅ Peptidoglycan cell wall support
🔗 github.com/pstansfeld/mempro
📖 pubs.acs.org/doi/10.1021/...

All I want for Christmas is your feedback in our preprint!
Happy to share work from Daniil Lepikhov, Laura Sandner and me, where we show that including information from the enzyme-substrate interaction interface in deep learning models can improve the prediction of Michaelis constants. #compchem
Take advantage of this fantastic opportunity to become part of the SCALE community! Several professorships and group leader positions are available at Goethe University and FIAS. We are looking for professors in the fields of molecular microbiology, cellular biochemistry, and molecular biochemistry.
19.12.2025 22:44 — 👍 18 🔁 16 💬 1 📌 0
picture of a woman in a plaid suit, red shirt, black pants, leaning on an industrial looking object and smiling at the camera

pictture of an old looking city in europe called freiburg germany, the picture prominently features an old catholic cathedral called the freiburg münster with hills in the background and dusky pink/blue sky

picture of a woman scuba diving on a shipwreck

picture of a woman in a halloween costume, the halloween costume is of the amino acid cysteine
Hi all!! At some point I passed 1K followers! Time for a re-intro! My name is Fiona! I am originally from Florida, did my postdoc at UCSD now I am a Junior Professor at the University of Freiburg in the Institute for Physical Chemistry, lets connect! I also love the ocean and halloween costumes!
13.12.2025 12:57 — 👍 34 🔁 10 💬 4 📌 1Exciting new preprint from PhD student Bryony Clifton in @adamgrieve.bsky.social lab with input from me, mixing AF2, BioEmu, and MD.
25.11.2025 12:57 — 👍 6 🔁 0 💬 0 📌 0
rdcu.be/eRxNf here our new study on the characterization of the lipopolysaccharide holotranslocon, LptDE and the newly identified subunits LptM and LptY. Great collaboration with @fronzeslab.bsky.social @pstansfeld.bsky.social @JulienMarcoux @YvesQuentin
24.11.2025 16:08 — 👍 31 🔁 15 💬 3 📌 1
Very happy to share our work on lipopolysaccharide assembly by the Lpt complex, published today in @natcomms.nature.com
Fantastic collaboration with @raffaeleieva.bsky.social l and t @pstansfeld.bsky.social
Congrats to all authors, especially Haoxiang, Axel & Violette
🔗 doi.org/10.1038/s414...
IT'S HAPPENING! 💥 I'm psyched to launch the collaboration between @qedscience.bsky.social & @openrxiv.bsky.social @biorxivpreprint.bsky.social! Preprint + q.e.d = your science is out there, and anyone can appreciate it. Let's care about making discoveries, and not on “getting published” (1/3) 👇
06.11.2025 14:49 — 👍 130 🔁 64 💬 7 📌 13Excited to share our latest @nature.com: How does naloxone (Narcan) stop an opioid overdose? We determined the first GDP-bound μ-opioid receptor–G protein structures and found naloxone traps a novel "latent” state, preventing GDP release and G protein activation.💊🧪 🧵👇 www.nature.com/articles/s41...
05.11.2025 16:22 — 👍 123 🔁 35 💬 6 📌 2
*If you would like to be considered for a talk at this workshop, please register & submit an abstract by 7 November*
We also have a fantastic line-up of invited speakers: Matteo Degiacomi, Lucie Delemotte, Weria Pezeshkian, Mohsen Sadeghi & Ilpo Vattulainen
Registration: aias.au.dk/events/show/...
🔬 CECAM Workshop: “Integrative and Multiscale Modelling Approaches to Illuminate CryoEM/ET Data”
Join us in Toulouse, 24–26 March 2026, to explore how molecular simulations, AI tools, can be integrated to cryo-EM and cryo-ET data.
🧬 Selection: brief CV + motivation letter required.
Apply 👇
📜 New preprint! We developed PoGö, an algorithm to optimize the essential dynamics of GöMartini proteins based on all-atom simulations: www.biorxiv.org/content/10.1...
27.10.2025 07:54 — 👍 33 🔁 12 💬 2 📌 4reach out to me on here or via my departmental email with any queries. To be cosupervised between myself and Prof Eamonn Kelly and Dr Alex Conibear.
24.10.2025 15:10 — 👍 0 🔁 0 💬 0 📌 0
Passionate about GPCRs and molecular pharmacology? We’re looking for a PDRA to use computational molecular modelling and in vitro approaches (BRET etc) to uncover how the nitazene classe of synthetic opioids work.
Learn more and apply: www.bristol.ac.uk/jobs/find/de...
Reposts appreciated!
Registration now open for the '3rd Annual Danish Workshop on Advanced Molecular Simulation'
Join us in beautiful Aarhus for a 2-day workshop on biomolecular simulations!
9-10 December 2025
Link for registration: aias.au.dk/events/show/...

Are the lipids associated with static protein structures there as long-lived ligands or an effect of preferential solvation? This computational-experimental framework shows the way! #lipidtime #compchem
www.nature.com/articles/s41...

Fancy doing a PhD in Bristol? Collab with us and @robincorey.bsky.social and Vicki Gold Labs.
Then check these out!
gw4biomed.ac.uk
www.findaphd.com/phds/project...

🚨🧠🔬 A major breakthrough in molecular neuroscience:
I am excited to share a new story from our lab, published in accelerated format today by @nature.com:
"Delta-type glutamate receptors are ligand-gated ion channels"
Read more here (free article link): rdcu.be/eGIKz
Academic authors, here's a peek into the black box of journal publishing from an journal editor if you can bear it:
06.09.2025 23:09 — 👍 1003 🔁 473 💬 18 📌 105Great opportunity for a recent PhD graduate to apply molecular dynamics and gain cryo-EM experience on an exciting project. Beautiful campus, world-class research institute, and a highly supportive, collaborative lab.
29.08.2025 12:29 — 👍 2 🔁 2 💬 0 📌 0
We are recruiting 2 postdocs to work on bacterial motility using a combination of cryoEM, FLM and MD. An exciting collaboration with Kirsty Wan @micromotility.bsky.social Wolfram Moebius @wolframmoebius.bsky.social and Daniel Kattnig. Please see links in the thread below!
28.08.2025 09:13 — 👍 27 🔁 15 💬 2 📌 2
Membrane Biology: Nothing can replace polyunsaturated lipids
Genetic studies reveal that polyunsaturated lipids do more than simply increase the fluidity of the cell membrane.
buff.ly/qcLJNUN

New from our lab!
🚀 ParametrizANI - a #NeuralNetworks tool for molecular parametrization.
Predicts potential energy surfaces with near-DFT/CC accuracy, at a fraction of the computational cost!
#AI #QuantumChemistry #CompChem #ML 🤖
chemrxiv.org/engage/chemr...
Try it:
github.com/palermolab/P...

👋🏻 #bluesky GPCR
🚀 Want to get to know me a bit better?
⚗️ Curious about what I’ve been working on?
I had a fun chatting on the Dr. GPCR #podcast to talk about my #journey, #research, and everything in between!
🎥 Watch on YouTube: lnkd.in/gzGQYX-x
🔗 Podcast: lnkd.in/gbcGj_HH
#GPCR #compchem
We are proud to be a founding contributor to lipidinteractome.org, a repository developed by @tafesselab.bsky.social & Schultz lab to increase accessibility to proteomics data from multi-functionalized lipid analogs! Check out the website & preprint: arxiv.org/abs/2507.23101 #lipidtime
05.08.2025 13:06 — 👍 62 🔁 19 💬 1 📌 1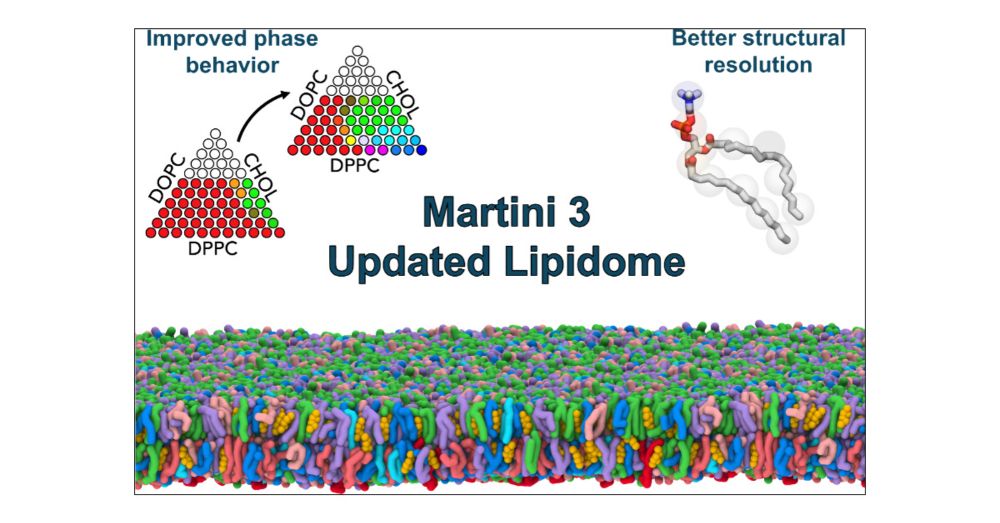
Our collaborative work "The #Martini3 #Lipidome: Expanded and Refined Parameters Improve Lipid Phase Behavior" is now published in #ACSCentralScience! 🎉
📄 Read: pubs.acs.org/doi/10.1021/...
💾 GitHub: github.com/Martini-Forc...
#MolecularDynamics #Biophysics #Simulations #Lipids

I’m really happy to share that I’ve passed my PhD viva in Computational Biochemistry! It’s been an exciting journey and I’m profoundly grateful to my supervisors Dr @marcvanderkamp.bsky.social and Dr Deborah Shoemark, and my collaborator Dr Richard Sessions for all their support and guidance.
25.07.2025 07:36 — 👍 6 🔁 1 💬 2 📌 0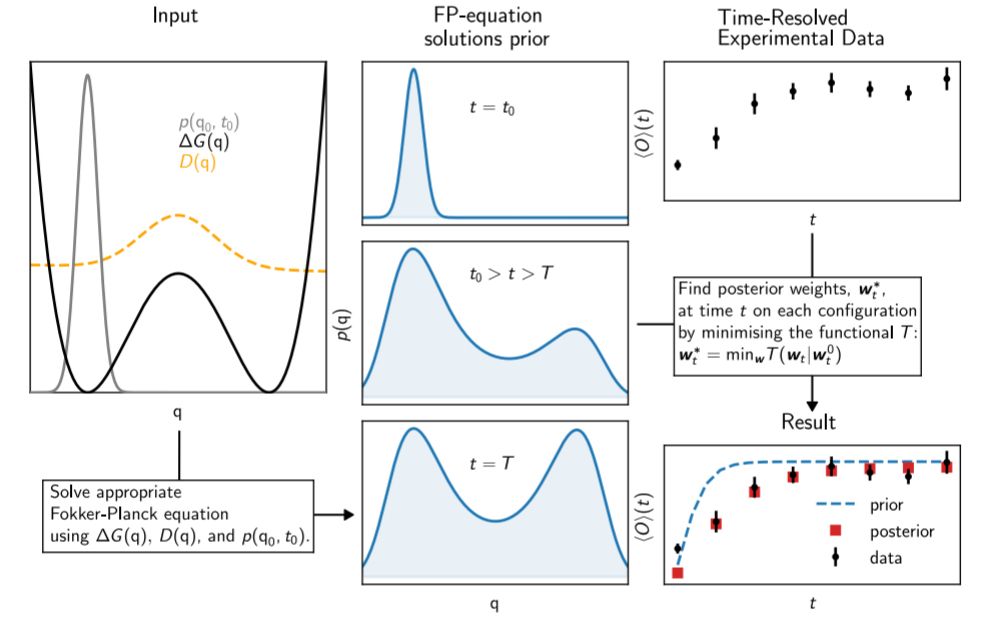
Once in a lifetime opportunity:
If you’re a bioRxiv screener and want to be the first to read
A Bayesian approach to interpret time-resolved experiments using molecular simulations
now is your chance 😉
Applications for this position are closing tomorrow! Get yours in quickly 😁
19.07.2025 02:03 — 👍 2 🔁 2 💬 0 📌 0