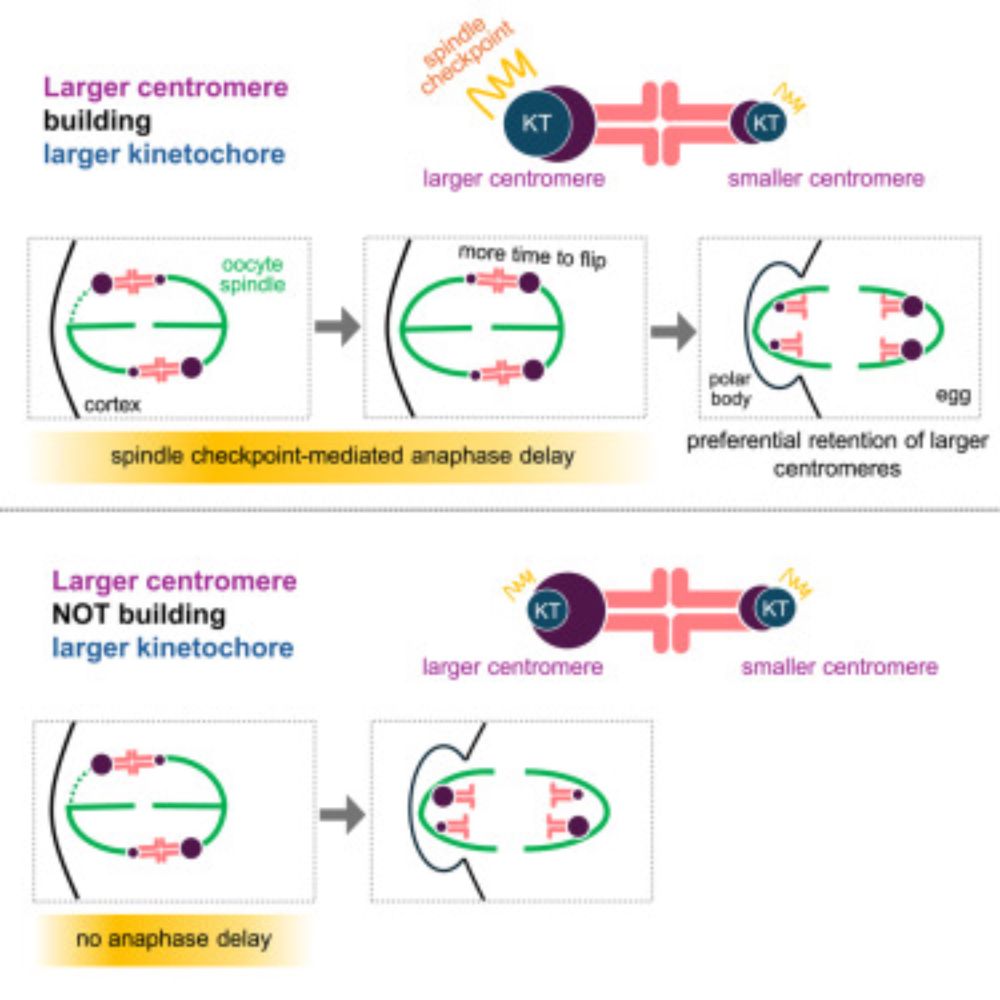
Spindle checkpoint can secure additional cheating time for selfish expanded centromeres cell.com/current-biol...
@currentbiology.bsky.social @takashiakeralab.bsky.social
@kerrycobb.bsky.social
Bioinformatics Analyst in the UConn Computational Biology Core. Affiliated with UConn Health.

Spindle checkpoint can secure additional cheating time for selfish expanded centromeres cell.com/current-biol...
@currentbiology.bsky.social @takashiakeralab.bsky.social
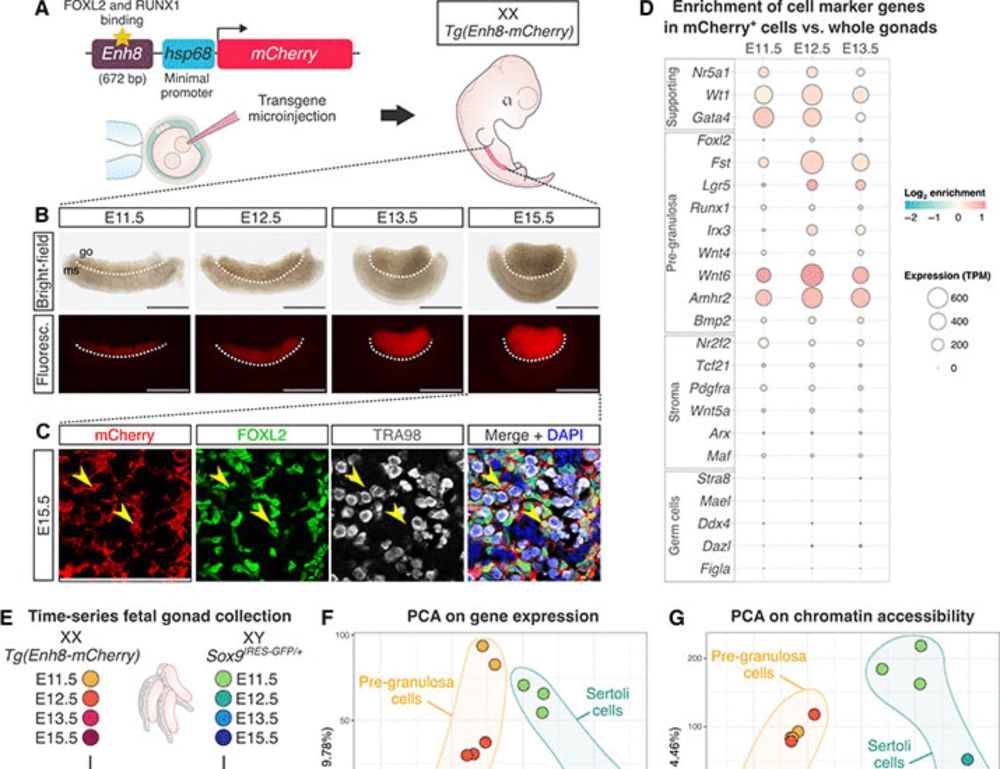
1/n Very excited to share our recent paper published on
@ScienceAdvances
where we dissect the gene regulatory landscape guiding mouse sex determination!
science.org/doi/10.1126/...
A huge well done to @IsabelleStevant.genomic.social.ap.brid.gy, Meshi and Elisheva who spearhead this project!
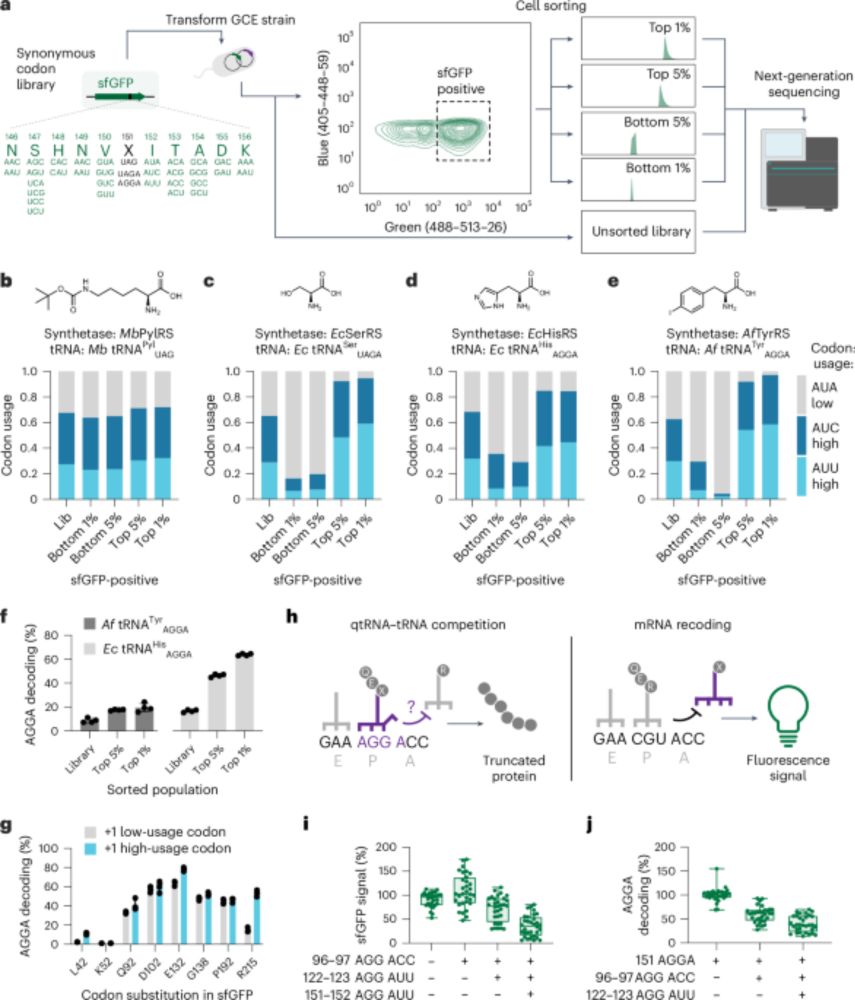
Noncanonical amino acids are efficiently incorporated into proteins by optimizing mRNA codon usage go.nature.com/3MDSSXQ
rdcu.be/exfiH
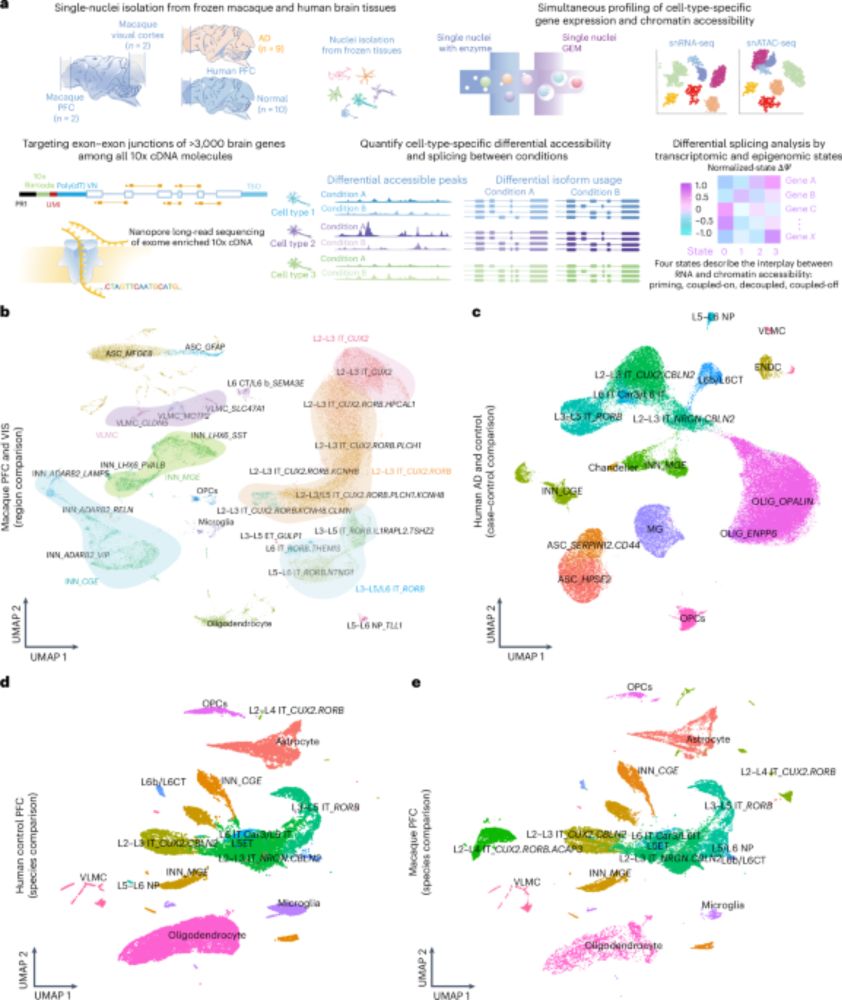
Combined single-cell profiling of chromatin–transcriptome and splicing across brain cell types, regions and disease state - @hagentilgner.bsky.social @liganlab.bsky.social go.nature.com/3IKlcJn
22.07.2025 14:43 — 👍 29 🔁 5 💬 0 📌 1
Fresh off the presses in @currentbiology.bsky.social : Is the deuterostome clade an artefact? www.sciencedirect.com/science/arti...
We (yours truly, Paschalis Natsidis, Laura Piovani and co-leads Paschalia Kapli and @maxjtelford.bsky.social) set out to try to answer this question.
Why? (1/12)

Benchmarking Imputed Low Coverage Genomes in a Human Population Genetics Context onlinelibrary.wiley.com/doi/10.1111/...
10.07.2025 07:54 — 👍 7 🔁 6 💬 0 📌 0Excited to launch our AlphaGenome API goo.gle/3ZPUeFX along with the preprint goo.gle/45AkUyc describing and evaluating our latest DNA sequence model powering the API. Looking forward to seeing how scientists use it! @googledeepmind
25.06.2025 14:29 — 👍 217 🔁 82 💬 5 📌 10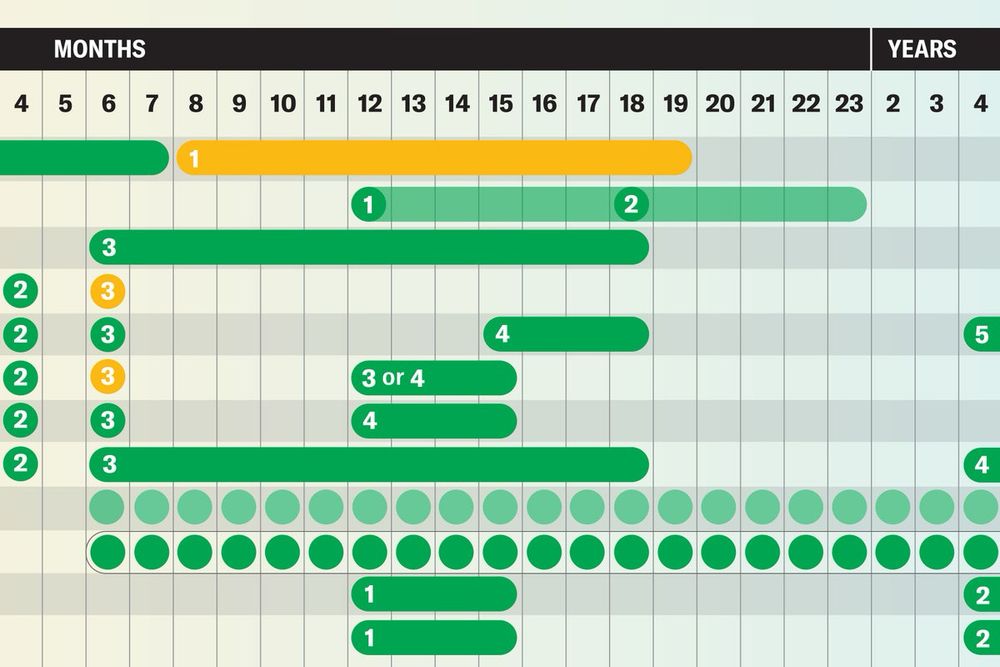
Vaccination schedules on the CDC website have already started changing under RFK Jr. So we published a guide to the evidence-based vaccine recommendations in place *before* all 17 members of the advisory panel were abruptly dismissed by the new admin. www.scientificamerican.com/article/see-... 🧪
25.06.2025 11:49 — 👍 10398 🔁 6132 💬 325 📌 286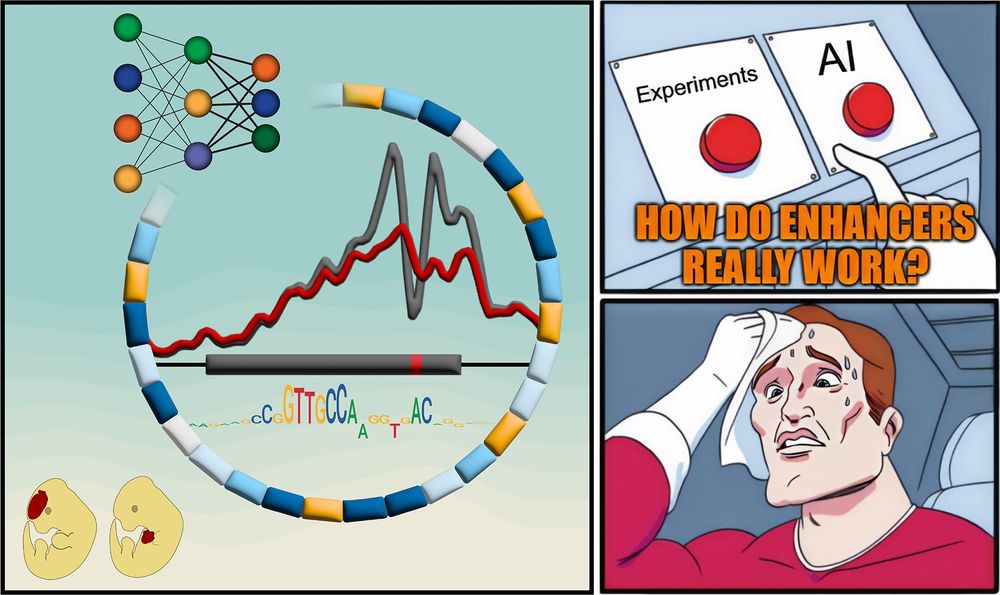
A meme-style comic panel with three parts. Left: A stylized enhancer with a mutation, surrounded by colored blocks representing functional motifs, a neural network diagram, chromatin accessibility signal traces, and a sequence motif. Two cartoon mouse embryos below show different LacZ reporter activity patterns. Top right: A hand hovers anxiously between two red buttons labeled “Experiments” and “AI,” with the caption “HOW DO ENHANCERS REALLY WORK?” Bottom right: A sweating superhero wipes his forehead, looking stressed about the difficult choice.
Textbooks: “Enhancers are just a bunch of TFBSs”
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22

Join us June 24–26 for a Virtual Single-Cell Transcriptomics Workshop! 🧬 Learn data processing, QC, clustering & integration using Python + R.
🕙 10 AM – 2 PM ET
💻 Zoom + recordings
💰 $500 ($400 UConn)
Register: bioinformatics.uconn.edu/cbc-workshops/
#scRNAseq #Bioinformatics #Workshop

Congrats to @dantipov.bsky.social et al. on the publication of Verkko2! The team put a ton of work into this making it the first assembler that deals with the complexity of human acrocentric chromosomes. Lots of interesting discoveries to come! genome.cshlp.org/content/earl...
17.06.2025 13:39 — 👍 32 🔁 19 💬 1 📌 1
I wrote a review of a recent paper on false discovery and multiple testing correction. liorpachter.wordpress.com/2025/06/16/r...
16.06.2025 21:31 — 👍 147 🔁 40 💬 7 📌 7
Preprint on "Improving spliced alignment by modeling splice sites with deep learning". It describes minisplice for modeling splice signals. Minimap2 and miniprot now optionally use the predicted scores to improve spliced alignment.
arxiv.org/abs/2506.12986
FYI: New online! RNA splicing — a central layer of gene regulation
11.06.2025 14:01 — 👍 17 🔁 3 💬 0 📌 0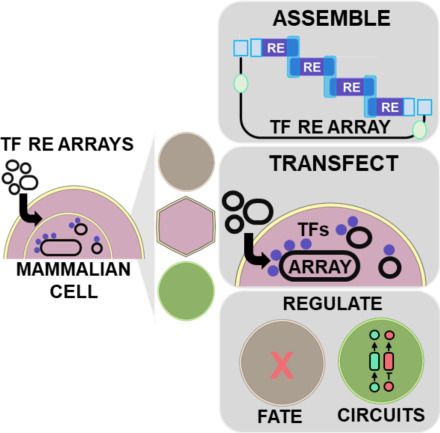
Annalise Zouein's main PhD research paper is published today in Trends in Biotech. Building and using repetitive transcription factor-binding arrays to modulate gene regulation in mammalian cells. No RNA, no protein - just DNA doing the work! www.sciencedirect.com/science/arti...
10.06.2025 12:33 — 👍 23 🔁 8 💬 1 📌 2
Today, we’re announcing the preview release of ty, an extremely fast type checker and language server for Python, written in Rust.
In early testing, it's 10x, 50x, even 100x faster than existing type checkers. (We've seen >600x speed-ups over Mypy in some real-world projects.)
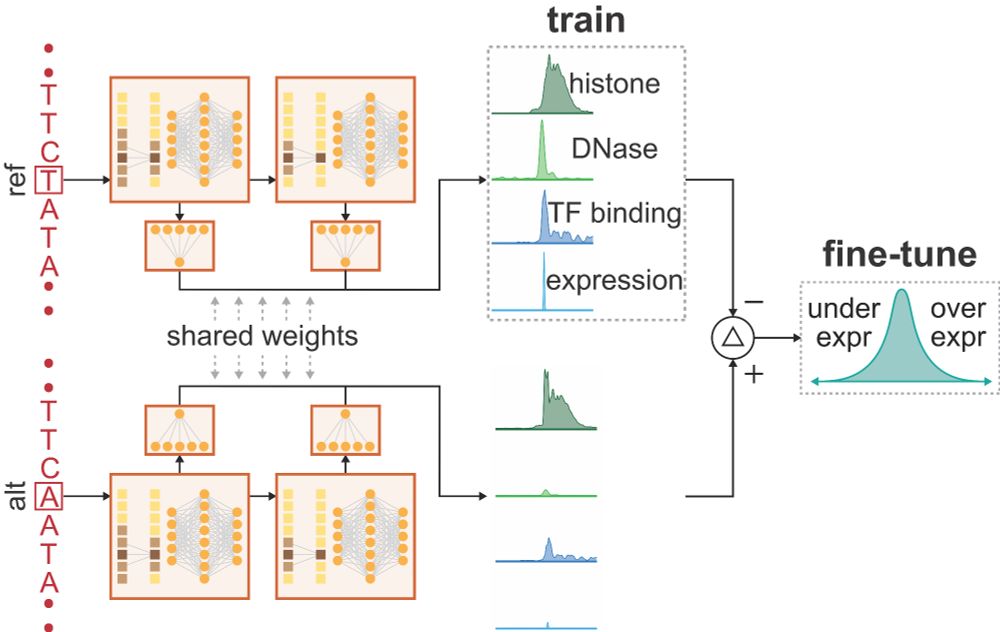
We're thrilled to introduce PromoterAI — a tool for accurately identifying promoter variants that impact gene expression. 🧵 (1/)
29.05.2025 18:29 — 👍 60 🔁 28 💬 1 📌 2
Expanding Perturb-seq-like screens to multicellular systems also increases the complexity of confounding effects. We encountered this when we perturbed all TFs in mosaic embryoid bodies (EBs) and develop a scalable solution by barcoding monoclonal individuals. 🧵 www.biorxiv.org/content/10.1...
25.05.2025 11:00 — 👍 62 🔁 14 💬 1 📌 2
How does tumour heterogeneity arise? How can we predict cancer cell plasticity? In 2 new studies, we trace #glioblastoma heterogeneity to a spatial cancer cell trajectory w. multimodal cell atlassing bit.ly/4mkrWgs & predict plasticity w. snRNA/ATAC+deep learning bit.ly/3FbI6Ic 🧵
16.05.2025 11:42 — 👍 71 🔁 31 💬 6 📌 5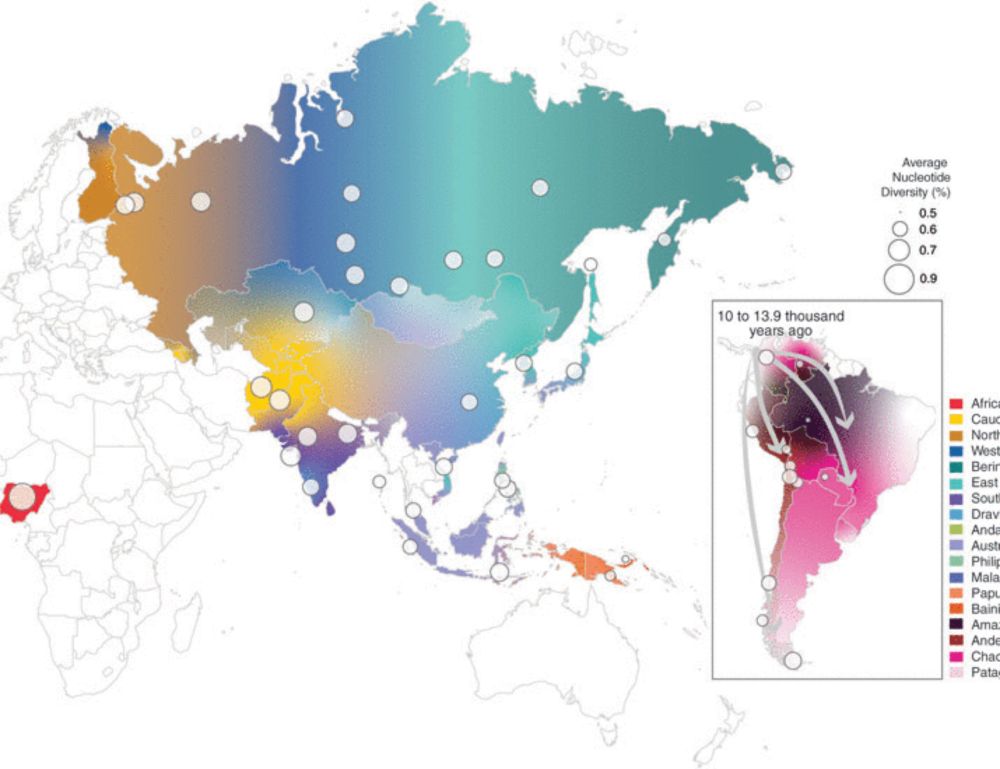
A large-scale genomic study in Science of over 1500 individuals from 139 underrepresented Indigenous groups across northern Eurasia and the Americas sheds new light on the ancient migrations that shaped the genetic landscape of North and South America.
Learn more: scim.ag/4mi1yUf

Spaces are still available in our upcoming genomics workshops! Join us for Variant Detection: May 20–22 and/or Genome Annotation, May 27-29 - Participants receive guided tutorials, live instruction, & daily recordings to support continued learning. bioinformatics.uconn.edu/cbc-workshops/
15.05.2025 12:59 — 👍 0 🔁 2 💬 0 📌 0
Here it is! Bonsai. Now there is really no more excuse for using t-SNE/UMAP. Bonsai not only makes cool pictures of your data. It actually rigorously preserves its structure. No tunable parameters. Incredible work by @dhdegroot.bsky.social.
I'm so excited about this!
www.biorxiv.org/content/10.1...
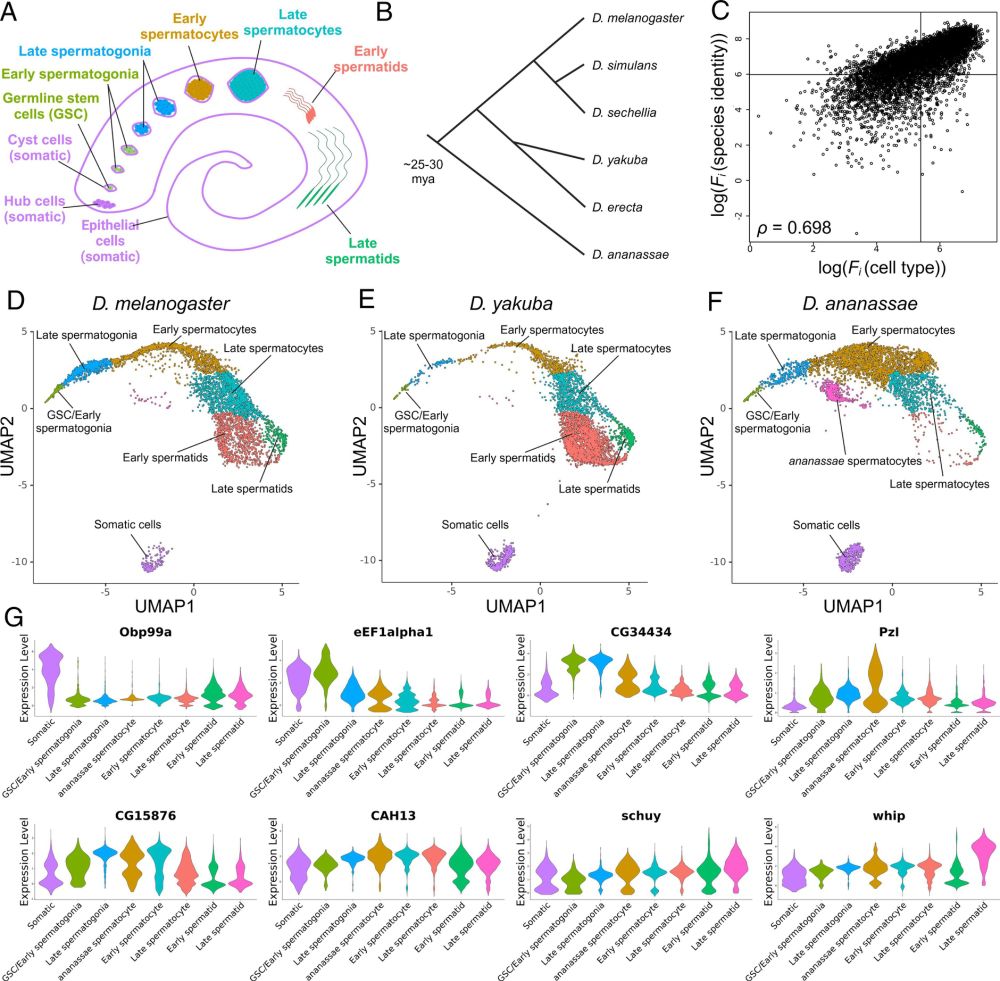
I’m very glad to share our recent publication
@pnas.org
, led by UnJin Lee from our lab at
@rockefelleruniv.bsky.social
: pnas.org/doi/10.1073/.... In this work, he 1) developed a method to efficiently compare single-cell RNA-seq data from rapidly evolving tissues across multiple species, and
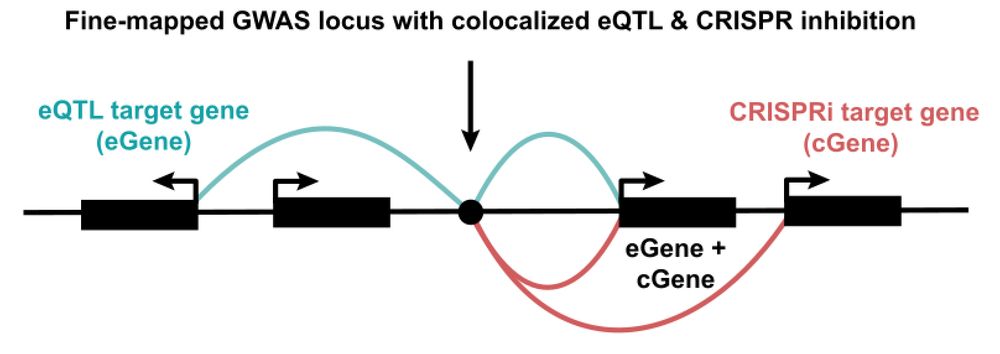
Our new contribution to the quest to find causal GWAS genes! Sam Ghatan from my lab at @nygenome.org led a systematic comparison of eQTLs and CRISPRi+scRNA-seq screens. TL;DR: they provide highly complementary insights, with ortogonal pros and cons. 🧵👇
www.biorxiv.org/content/10.1...
Mapping the gene regulatory landscape of archaic hominin introgression in modern Papuans https://www.biorxiv.org/content/10.1101/2025.05.04.652069v1
05.05.2025 02:31 — 👍 11 🔁 4 💬 0 📌 2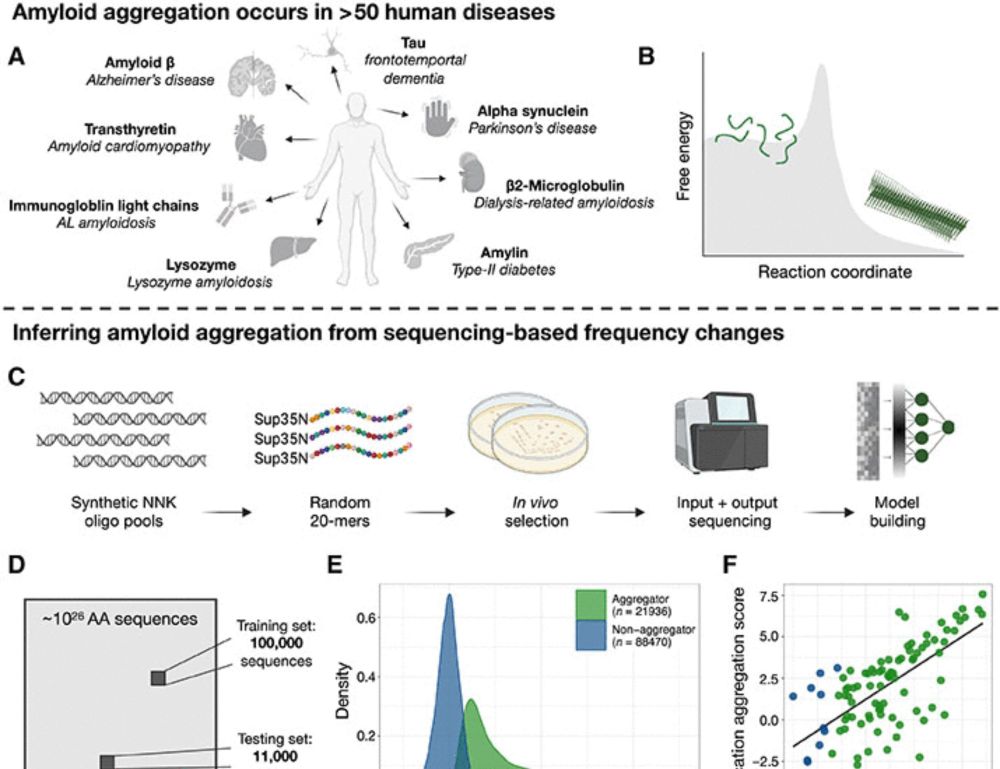
We quantify the aggregation of >100,000 random protein sequences to train CANYA, a convolution-attention hybrid neural network to predict aggregation from sequence. With @bennibolo.bsky.social www.science.org/doi/10.1126/...
04.05.2025 10:40 — 👍 77 🔁 31 💬 1 📌 2Check out this great resource of single cell multiome data, cell annotations, regulatory DNA models of cell type resolved chromatin accessibility & comprehensive annotation of motifs & sequence syntax for multiple tissues in early human development 1/
04.05.2025 01:43 — 👍 72 🔁 14 💬 3 📌 0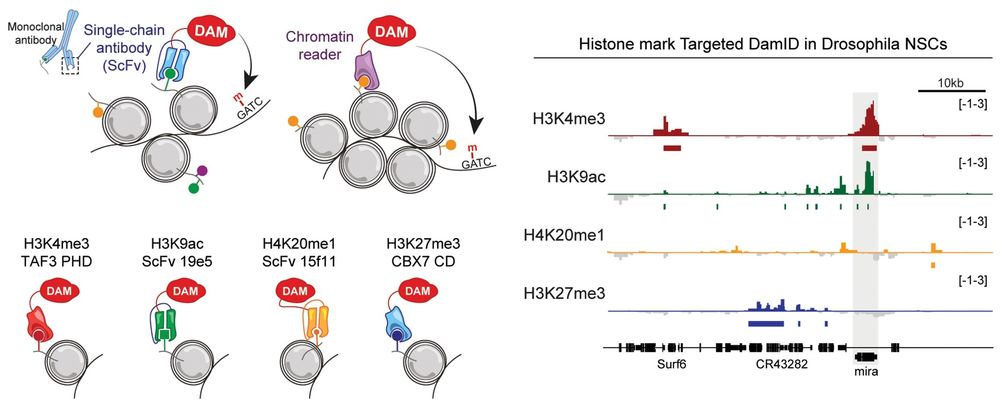
Profiling histone marks with TaDa. Top left: Schematic showing methods used for Targeted DamID profiling of histone modifications. Bottom left: Illustration demonstrating fusion of Dam to chromatin-binding domains for H3K4me3 or H3K27me3 and to mintbodies (scFvs) against H3K9ac or H4K20me1. Right: Histone marks near the Mira locus (shaded) in Drosophila third-instar neural stem cells.
Current methods for profiling #HistoneModifications require cell isolation & don't work with all tissue types. @jellevda.bsky.social &co adapt Targeted DamID (TaDa) to profile cell-type-specific #histone marks throughout the genome, in vivo, without cell isolation 🧪 @plosbiology.org plos.io/3Dw8sUD
30.04.2025 10:45 — 👍 21 🔁 11 💬 0 📌 0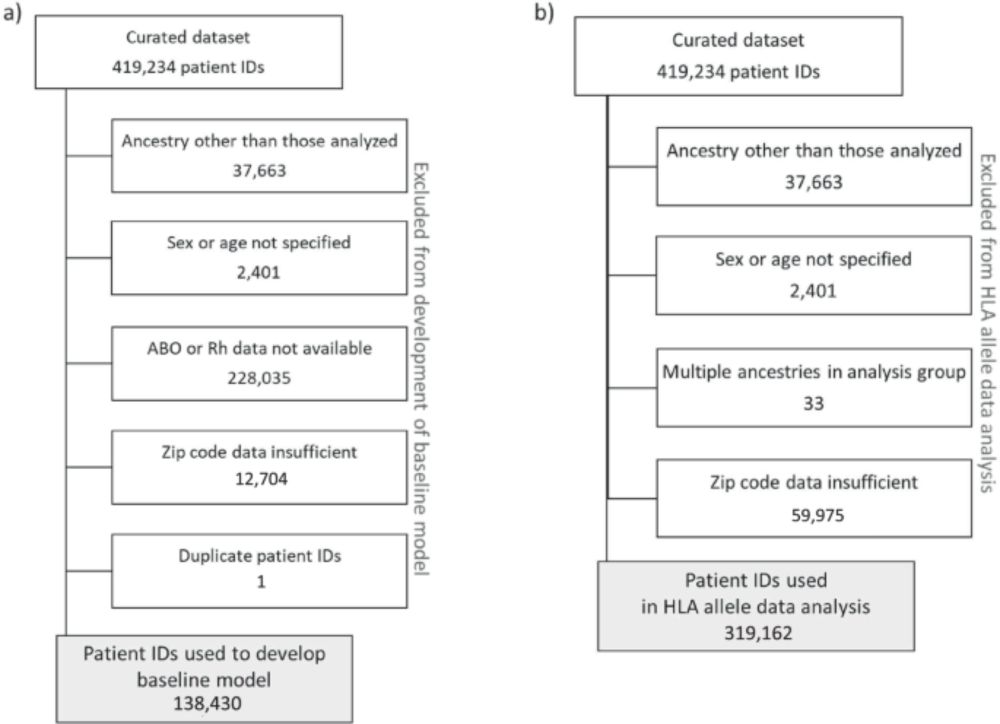
🚨 Association between #HLA #genetics and #SARS-CoV-2 #infection in a large #real-#world #cohort
👉 #genes & #immunity @springernature.com
www.nature.com/articles/s41...

Excited to share the first manuscript from my PhD in which we leveraged ultra-long Nanopore sequencing, D. melanogaster inbred lines, and a ton of manual validation to investigate the effects of long-read length on population-level structural variant (SV) calling accuracy! doi.org/10.1101/2025...
25.04.2025 20:03 — 👍 55 🔁 24 💬 1 📌 1