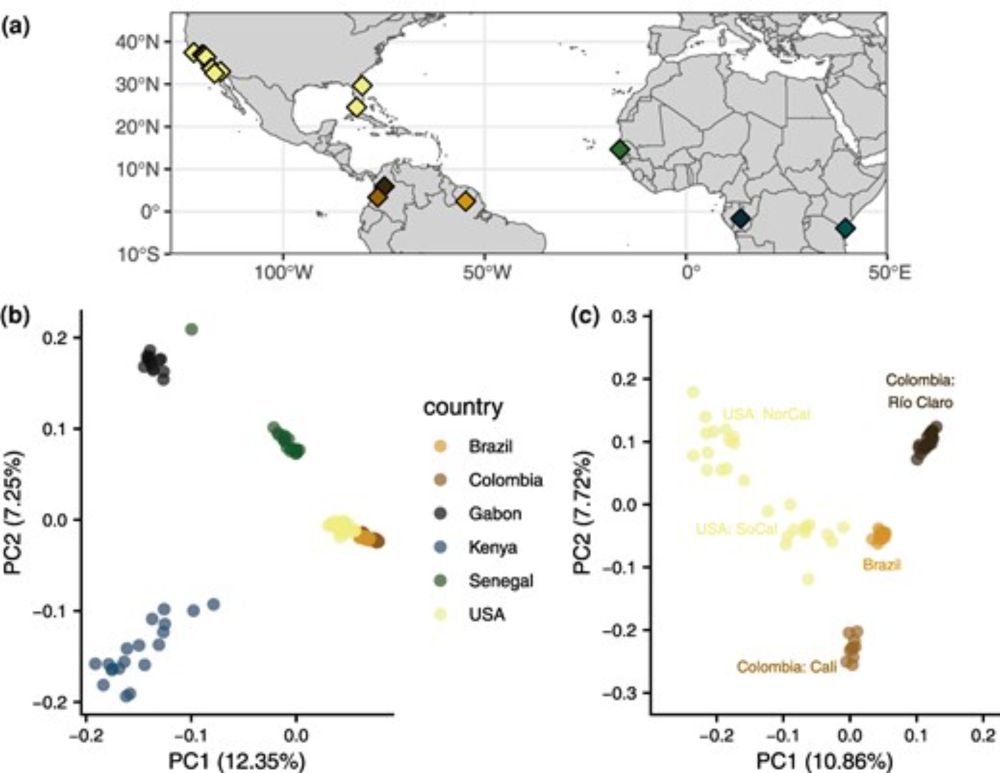
Demographic History, Genetic Load, and the Efficacy of Selection in the Globally Invasive Mosquito Aedes aegypti
Abstract. Aedes aegypti is the main vector species of yellow fever, dengue, Zika, and chikungunya. The species is originally from Africa but has experience
New research by
@tylervkent.bsky.social @samurscicop.bsky.social & D. Matute analysed 131 genomes of the globally invasive Aedes aegypti, and found high genomic resilience despite historical eradication attempts for this globally invasive vector.
🔗 doi.org/10.1093/gbe/...
#genome #mosquitoes
29.04.2025 14:36 — 👍 19 🔁 10 💬 0 📌 0

Demographic history, genetic load, and the efficacy of selection in the globally invasive mosquito Aedes aegypti
Abstract. Aedes aegypti is the main vector species of yellow fever, dengue, Zika and chikungunya. The species is originally from Africa but has experienced
Excited to see the final version of this paper with @samurscicop.bsky.social and Daniel Matute out in @genomebiolevol.bsky.social!
We took a dive into the complicated demographic history of Aedes aegypti mosquitoes and its impact on the distribution of genetic diversity
doi.org/10.1093/gbe/...
07.04.2025 19:53 — 👍 49 🔁 24 💬 4 📌 1
YouTube video by Ryan Gutenkunst
Talk describing the results of GHIST1 at ProbGen 25.
Amid the chaos, it was great to share results from the first Genomic History Inference Strategies Tournament at ProbGen25. Watch the talk to learn about the future of GHIST and @andrewhvaughn.bsky.social's nefarious metagaming of demographic inference. 😀 www.youtube.com/watch?v=3_dv...
17.03.2025 15:53 — 👍 19 🔁 12 💬 1 📌 0
These results highlight a key challenge for population-genetic analyses in highly selfing species or low-recombination genomic regions. Check out the paper for a deeper dive into other potentially relevant factors like beneficial mutations, dominance coefficients, and population structure!
05.03.2025 18:36 — 👍 0 🔁 0 💬 0 📌 0
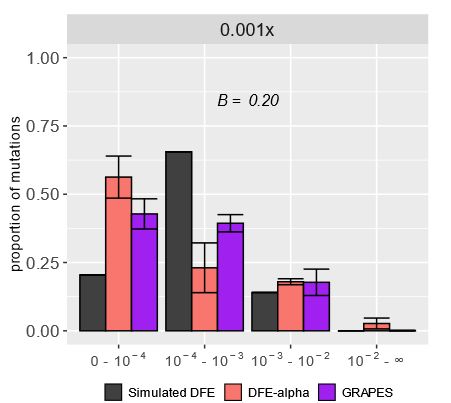
If HRI was the cause mis-inference, we’d expect it to be caused by the reduced levels of effective recombination in selfers. Indeed, when we ran simulations with lower recombination (but no selfing), we saw the same patterns of DFE mis-inference.
05.03.2025 18:36 — 👍 0 🔁 0 💬 1 📌 0

We hypothesized this mis-inference was caused by HRI, where linked deleterious mutations interact and reduce the efficacy of selection. The site frequency spectrum (SFS) had a U-shape at high selfing rates, a pattern often linked to HRI and not modeled by current DFE inference approaches.
05.03.2025 18:36 — 👍 0 🔁 0 💬 1 📌 0
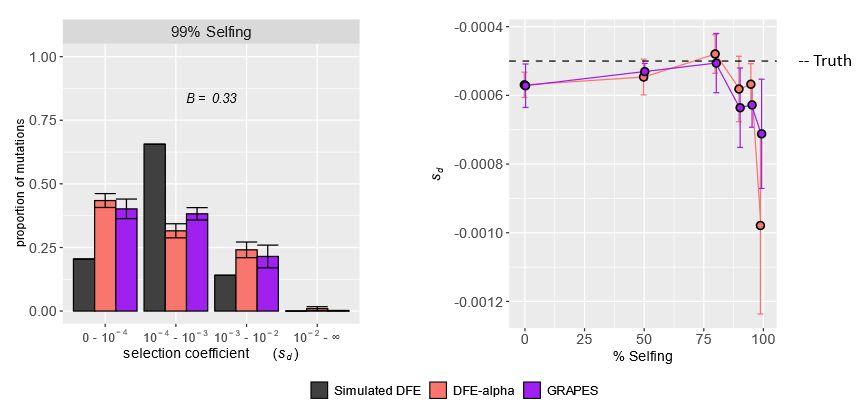
In simulated highly selfing populations, the DFE was mis-inferred by two unique DFE inference methods—nearly neutral and strongly deleterious mutations were overestimated, while mildly deleterious ones were underestimated.
05.03.2025 18:36 — 👍 0 🔁 0 💬 1 📌 0

Excited to share my first PhD project with my mentor, @johriparul.bsky.social! We examine how Hill–Robertson interference (HRI) in highly selfing species biases estimates of the distribution of fitness effects of new mutations (DFE).
doi.org/10.1093/evol...
@journal-evo.bsky.social #popgen #evobio
05.03.2025 18:36 — 👍 21 🔁 9 💬 1 📌 0
Thrilled to have been part of the inaugural GHIST competition in population genetics inference! Big thanks to the organizers for a fun and challenging event. Congrats to Andrew Vaughn and Ekaterina Noskova for their impressive performance, I'm looking forward to reading about everyone's methods.
11.12.2024 16:46 — 👍 0 🔁 1 💬 0 📌 0
Same content, different website! Human evolutionary genomics, functional genomics and archaic hominins. Group leader in Human Genomics at SVI in Melbourne/Naarm, Australia. Sometimes I go to Estonia.
Reimagining Science.
sciforgood.org
Assistant Member and Principal Investigator
Donald Danforth Plant Science Center, St. Louis MO
Interested in plants, popgen, polyploidy, and puns.
PhD Student at UMich Statistics.
The account mostly trashes about urban planning and infrastructure.
Probability, Statistics, and Evolutionary Biology.
https://hanbin973.github.io
Bioinformatics @urgi-inrae.bsky.social
Interested in #Genomics, #Evolution, #Transposons, Repeat annotation, #Pangenomic, #KnowledgeGraph
https://eng-urgi.versailles-grignon.hub.inrae.fr/about-us/team/johann-confais
join the TEhub : @tehub.bsky.social
PhD (Math Bio) @ Max Planck EvolBio. BS-MS (Physics) @ IISc Bangalore. Complex dynamical systems ⚙️
Bookworm, coder, gamer and pianist
Website: chanrt.github.io
Statistical geneticist. Professor of Human Genetics and Biostatistics at the University of Pittsburgh. Assiduously meticulous.
Faculty Penn State Bio. Director Penn State Herbarium. He/him.
laskylab.org
https://www.huck.psu.edu/institutes-and-centers/plant-institute/herbarium/about-the-herbarium
Professor passionate about student success & team science #firstgen #Brassica enthusiast walking Dogs of the Plant World
#Agriculture #Food #HigherEd #EduSky #Agsky #SciComm & more
#Soil #Crop #Science ; opinions mine
PhD student in the Samuk Lab at UC Riverside. Interested in evolutionary genetics
⊹ ࣪ ˖ 𓆦 ♡ 𓆝 ⊹ ࣪ ˖
Evolutionary geneticist studying recombination, hybridization, and domestication in yeast! Asst. Professor at NC State
🔬🧬🍞
👦👦🐈🐈
she/her
Evolutionary ecologist, Botanist, Associate Professor at Davidson College (PUI 💪🏼), she/her
🔗smwadgymar.weebly.com
Group Leader @GReD_Clermont
Chargé de Recherche @INSERM
Evolutionary biologist studying chromatin also drummer.
Jazz and epistemology. he/his
https://www.igred.fr/en/team/evolutionary-epigenomics-and-genetic-conflicts/
Evolutionary biologist, science writer, YouTuber. Assistant Professor at Augusta University. he/him 🏳️🌈
http://www.youtube.com/@talkpopgen
Amateur Scientist lost in genome space. Experimental evol. using Truncating ECAs.Polynesian sailing canoe enthusiast. Enjoy doing science for fun, really hate writing. All my views are temporary and subject to change without notification.
Assistant Professor at UNC Chapel Hill
Population genetics | Uppsala
Research Associate at University of Maine. Cultural Evolution, Climate Change Adaptation, Contaminated Land Cleanup
doctoral researcher
experiments on cooperation
Opinions expressed are my own.
Website: https://adileyasar.github.io
I study moths so YOU don't have to. Lepidopterist at Carnegie Museum of Natural History, Pittsburgh, PA, U.S.A. All opinions are my own.








