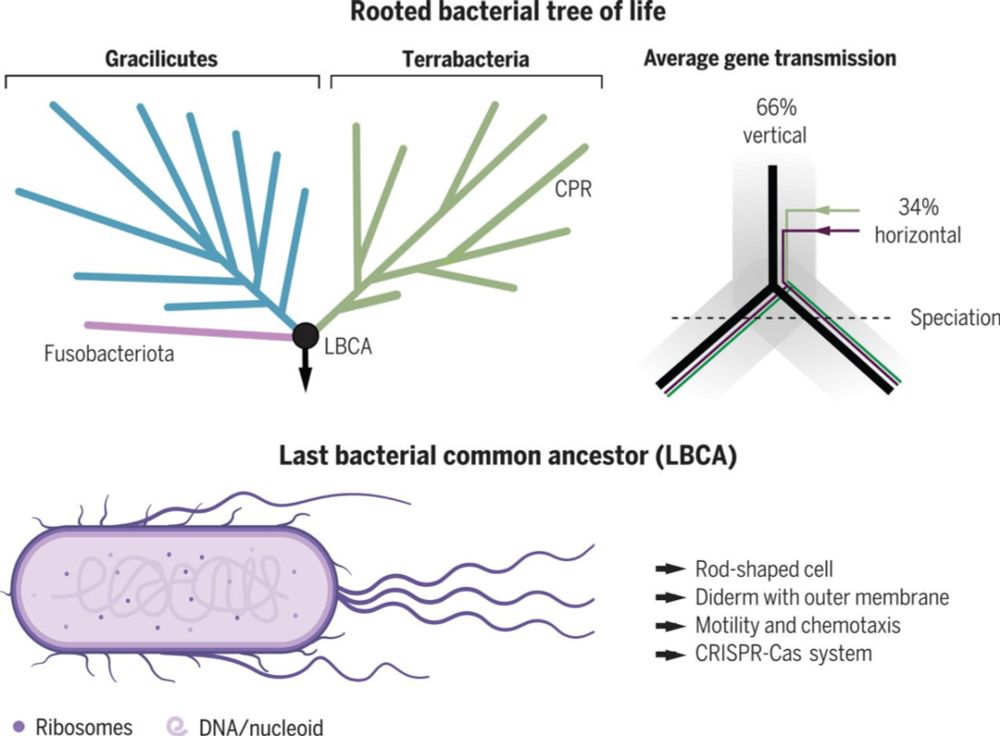
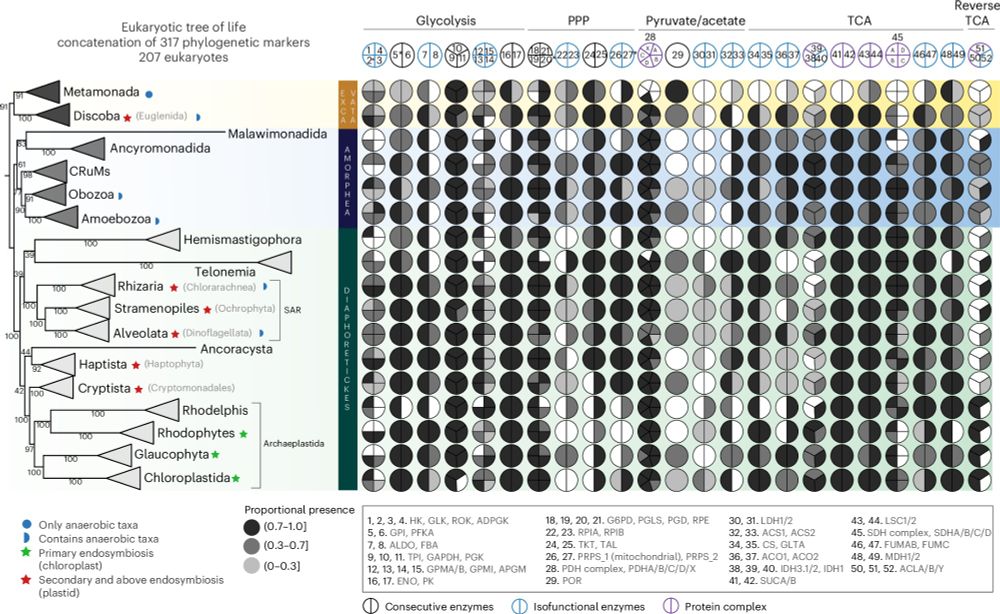
Our analyses converge on a narrow archaeal root region at/near the base of the Euryarchaeota, supporting hypotheses in which the Last Archaeal Common Ancestor was a complex, free-living (hyper-)thermophilic methanogen.
13.11.2025 19:28 — 👍 6 🔁 3 💬 1 📌 1
I'm looking for a PhD student to dive into some protist genomes and maybe find some cool parasites like this (very flexible) apicomplexan.
www.findaphd.com/phds/project...
Please do reach out if you're interested. And APPLY EARLY! The advert will be taken down when a suitable candidate is found.🪱🦀🐟
09.10.2025 16:02 — 👍 10 🔁 7 💬 1 📌 0

The genomic basis of symbiotic integration at University of Bath on FindAPhD.com
PhD Project - The genomic basis of symbiotic integration at University of Bath, listed on FindAPhD.com
There's a PhD position now available with me in Bath, on the evolution of symbiosis. www.findaphd.com/phds/project.... The supervisory team also includes @anja1.bsky.social @phil-donoghue.bsky.social and others. NB, this is open both to UK-based students *and* to international students :)
09.10.2025 09:39 — 👍 26 🔁 32 💬 0 📌 1
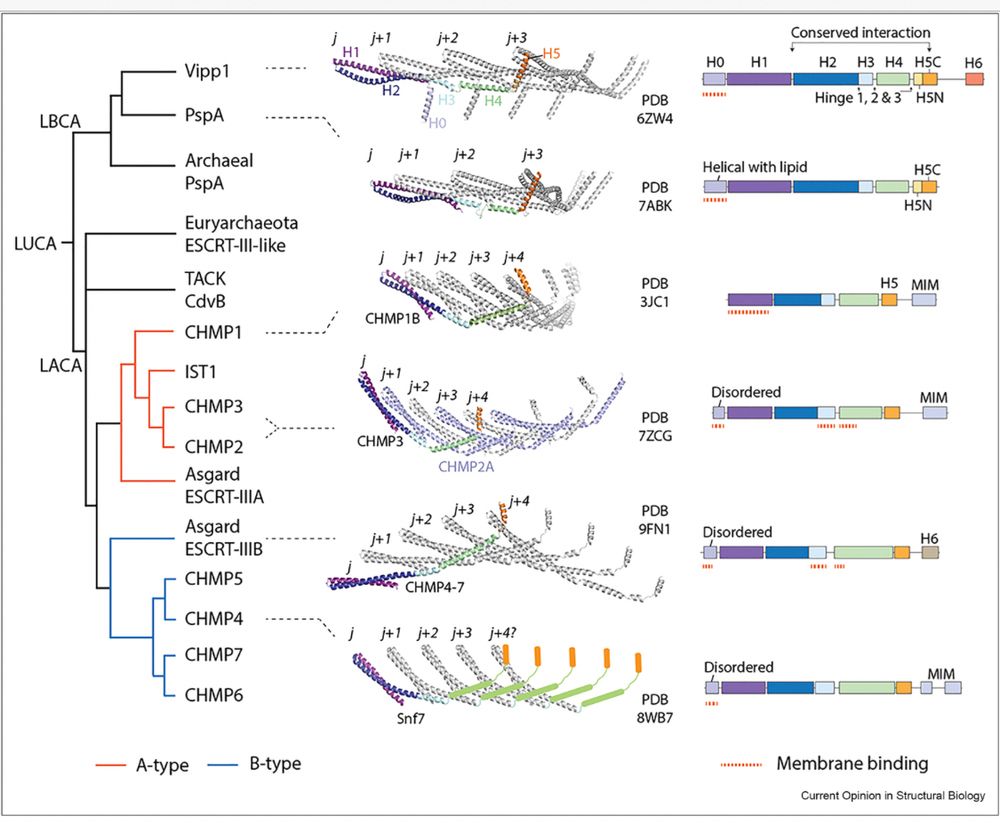
❗️ Exciting review alert❗️focussing on the evolution and mechanism of prokaryotic ESCRT-III-like systems.. fun collaboration with Tom Williams @tweethinking.bsky.social!
doi.org/10.1016/j.sb...
14.07.2025 17:19 — 👍 11 🔁 1 💬 0 📌 0
17.06.2025 13:09 — 👍 1 🔁 0 💬 0 📌 0
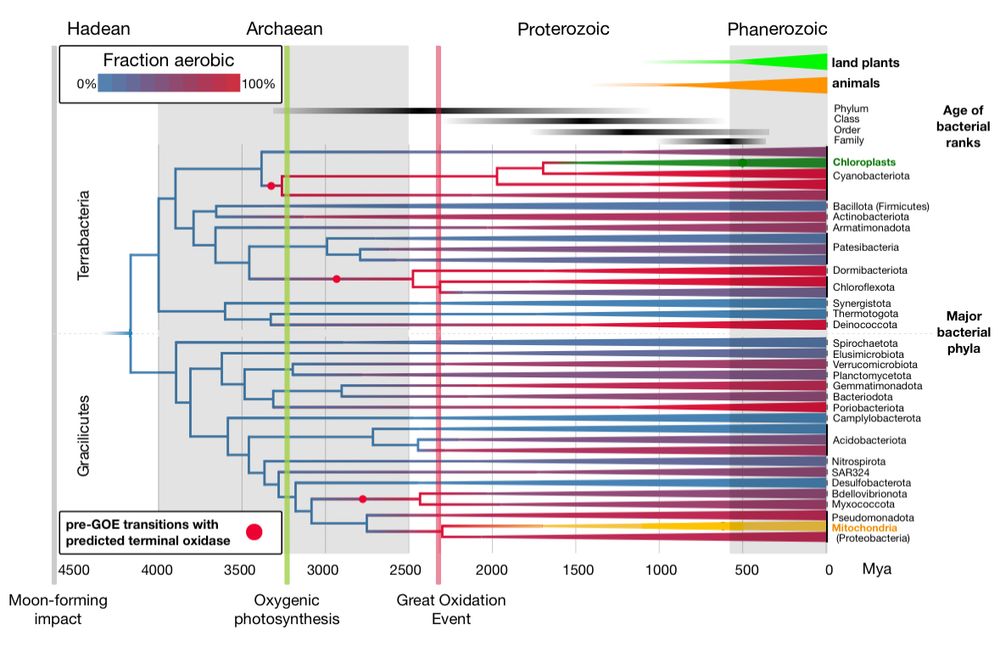
Dating Bacteria is hard due to the lack of maxima. Assuming aerobes likely postdated the GOE gave us better resolved ages, but also surprised us, but not Dr Dayhoff, showing O2 use predated its atmospheric rise by 900 Mys and helped oxygenic photosynthesis to evolve. www.science.org/doi/10.1126/...
04.04.2025 01:20 — 👍 43 🔁 26 💬 8 📌 4
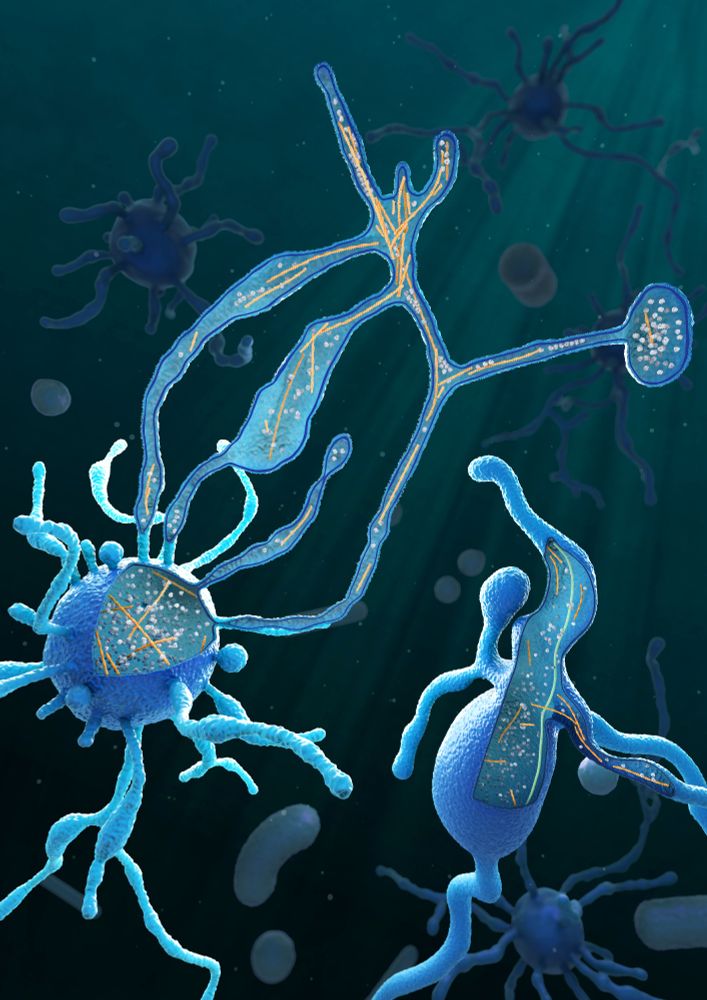
Credit: Margot Riggi (@margotriggi.bsky.social), Max-Planck Institute of Biochemistry (@mpibiochem.bsky.social)
Asgard archaea have actin - but what about microtubules? Where do they come from? 🧐 Our new paper www.cell.com/cell/fulltex... by @xujwet.bsky.social & @florianwollweber.bsky.social, in collaboration with the Schleper & Wieczorek labs, describes tiny Asgard microtubules! #TeamTomo #ArchaeaSky 1/6
21.03.2025 15:16 — 👍 256 🔁 110 💬 7 📌 15
Very interesting indeed for enthusiasts of symbiotic integration...
20.03.2025 19:49 — 👍 1 🔁 0 💬 0 📌 0
Project summary | rediploidisation.org
The position will involve developing pipelines and collaborating with other postdocs working on specific case studies of WGD across these different locations. See project website here www.rediploidisation.org. Feel free to contact James (jc493@bath.ac.uk) or me for inquiries, or see the ad.
18.02.2025 09:26 — 👍 0 🔁 0 💬 0 📌 0
The position will focus on phylogenomic and comparative genomic analyses of whole genome duplication and asynchronous rediploidization in eukaryotes. It's part of a wider large-scale grant (BBSRC sLOLA), with partners across the UK and further afield (e.g. Edinburgh, Oxford, Kew, Bristol, TCD, UCD).
18.02.2025 09:26 — 👍 0 🔁 0 💬 1 📌 0
ED12442 Research Associate in Phylogenomics (fixed-term) - Jobs at Bath
Job announcement: a four-year postdoc position in phylogenomics at the Milner Centre for Evolution, Bath, with James Clark and me (I'm moving to Bath later in the year...). www.bath.ac.uk/jobs/Vacancy...
18.02.2025 09:26 — 👍 8 🔁 7 💬 2 📌 0
Tagging @anja1.bsky.social and @wentsunghwang.bsky.social 🤣
17.02.2025 17:09 — 👍 2 🔁 0 💬 0 📌 0
These are Asgard archaea that have evolved by genome reduction from a more complex ancestor, perhaps due to evolving a symbiotic lifestyle … not the only Asgards that have trodden that path…
17.02.2025 16:59 — 👍 0 🔁 0 💬 1 📌 0
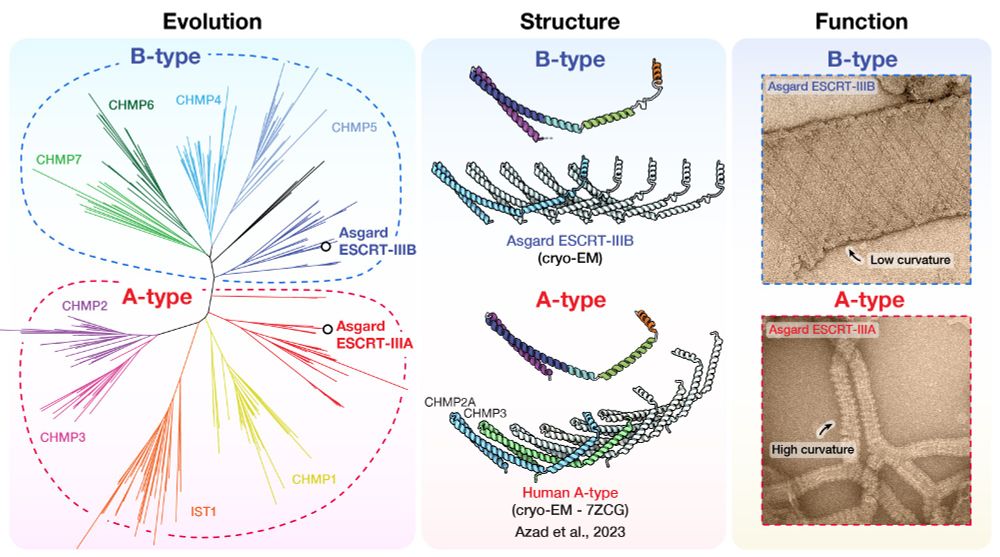
Evolution, structure and membrane remodelling function of the ESCRT-III superfamily in eukaryotes and their closest relatives Asgard archaea. The two subfamilies, B-type and A-type, have distinct structural properties to perform sequential steps of the membrane remodelling pathway.
New research on ancient Asgard archaeal ESCRT-III proteins reveals evolutionary secrets of membrane remodelling.
Diorge Souza, Javier Espadas and Sami Chaaban, investigated the ESCRT-III proteins with Buzz Baum and Aurelien Roux.
Read more: www2.mrc-lmb.cam.ac.uk/asgard-archa...
#LMBResearch (1/2)
11.02.2025 12:46 — 👍 33 🔁 12 💬 1 📌 1
Nice to see this out :)
08.02.2025 09:43 — 👍 4 🔁 0 💬 0 📌 0
Yay! Our LUCA study shorturl.at/09YR8 made into @quantamagazine.bsky.social list of 2024's biggest breakthroughs! Thanks to @emoody.bsky.social @tweethinking.bsky.social Davide Pisani @bristolpalaeo.bsky.social and the rest of our amazing team!
19.12.2024 16:52 — 👍 6 🔁 2 💬 0 📌 0
Marine biologist, fish biologist, restoration ecologist, science communicator & ocean optimist I Professor of Marine Biology & Global Change, School of Biological Sciences, University of Bristol I Co-Director: MSc Science Communication for a Better Planet
PI and Lecturer, University of Manchester
https://dannagifford.com @MERManchester.bsky.social
Computational Biologist. Tenure Track Fellow in Health at the University of Liverpool. 🇧🇷🇯🇵🇪🇸🇬🇧🇨🇭
Phylogenomics and Chemometrics. Prog Rock and Post-Punk. C and Python.
@leomrtns@mstdn.science
Theoretical biologist and advisor to data scientists at the University of Arizona. Mostly theoretical population genetics and molecular evolution, but I've also published in biochemistry, infectious disease, aging, economics, education. Opinions are my own
Career Development Research Fellow at St John's College, University of Oxford.
Evolution, comparative genomics, cooperation, horizontal gene transfer, plasmids.
https://www.anna-dewar.com/
How cells stick to things and move around, microscopy, cats, garden bugs, forays into machine learning, occasional political snark. Asst Prof University of Bath, UK 🏳️🌈
Theoretical Physicist.
(She/her/hers)
From Eastern KY.
Okinawa Institute of Science and Technology: https://www.oist.jp
沖縄科学技術大学院大学: https://www.oist.jp/ja
Algae enthusiast, Associate Professor @UniofExeter, joint appointment @thembauk
Algal ecophysiology | signalling | microbiome | molecular microbiology | diatoms!
Lecturer in Microbiome & Health at @apcmicrobiomeirel.bsky.social & @ucc.bsky.social
Alumnus @borklab.bsky.social
Microbiome, microbial ecology & metagenomics.
Computational and evolutionary biologist, studying virus, microbial and cancer evolution. Views my own.
Protein structure evolution | Royal Society Newton International Fellow at UCL | she/her
Protein evolution, phylogenetics, Tree of Life
PostDoc
Associate Professor, Inst. Plant & Microbial Biology, Taipei, Taiwan
Adjunct Faculty, National Taiwan University
EMBO Global Investigator
#Evolution #Eukaryote #Algae #GiantVirus #Genomics
#Microbes in our environment
https://chuanku-lab.github.io/kulab
Computational microbiologist
I like to post about: microbial genomics, microbial ecology, evolution, micro+plant biotechnology, climate, symbiosis, virology, ag, sci publishing and policy
I do science with fossils, X-rays and computers at the Unviersity of Manchester and Natural History Museum, UK. Working at Museum für Naturkunde, Berlin until September '25. Run the Palaeontological Association web systems.
russellgarwood.co.uk
Group leader at MEDEA Lab (Microbial Ecology, Diversity, Evolution, and Astrobiology), Kiel University .
Wildlife, micro-, macro-, astrophotography. Vandal of stringed instruments.
Opinions are my own. (he/him)
Evangelist for the study of evolution in action by everyone.
Pitt Prof | EvolvingSTEM | biofilms | EvMed | genomics entrepreneur (@SeqCoast.bsky.social, @midauthorbio.bsky.social) | ASM President-Elect | exercise addict ~ swim bike run
Microbial generalist 🦠 | Keen freediver & birdwatcher | Reluctant #Rstats enthusiast | she/her