
1H R1ρ Relaxation Identifies a Hidden Intermediate in DNA Base-Pairing
1H R1ρ Relaxation dispersion (RD) NMR experiments provide valuable atomic-level insights into transient, high-energy conformational states of biomolecules. However, cross-relaxation artifacts can hamp...
new #DNA #excitedstate just dropped💥 when my colleagues investigated how to avoid cross-relaxation artifacts in proton R1ρ experiments, which are used to measure #dynamics of #biomolecules, they discovered an excited state of DNA that may have a role in binding anticancer drugs
🔗 bit.ly/4o5m7U2
06.10.2025 18:47 — 👍 0 🔁 0 💬 0 📌 0
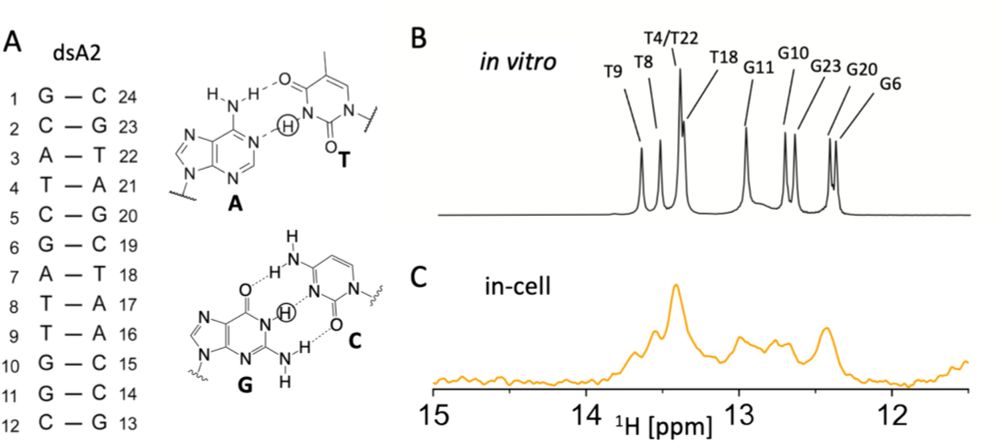
Optimising in-cell NMR acquisition for nucleic acids - Journal of Biomolecular NMR
Understanding the structure and function of nucleic acids in their native environment is crucial to structural biology and one focus of in-cell NMR spectroscopy. Many challenges hamper in-cell NMR in ...
Super cool #RNAdynamics have been discovered by NMR. Next, we want to observe these dynamics in their native environment with #in-cellNMR. Current limitations include low sensitivity. My colleagues help address this by measuring selective T1 to shorten acquisition time.
🔗 bit.ly/4doL8FD #scicommUU25
21.05.2025 12:37 — 👍 5 🔁 1 💬 0 📌 0

Base-pair conformational switch modulates miR-34a targeting of Sirt1 mRNA - Nature
Repression of a messenger RNA by a cognate microRNA depends not only on complementary base pairing, but also on the rearrangement of a single base pair, producing a conformation that fits be...
Here is a phenomenal example of why we care about #RNAdynamics. My group shows an excited state of a miRNA:mSirt1 complex is involved in downregulation of Sirt1, a tumour promoter. Identifying these excited states can help guide design of miRNA mimics to treat cancer.
🔗 bit.ly/4kcBKI4 #scicommUU25
21.05.2025 09:57 — 👍 1 🔁 0 💬 0 📌 0

RNA Dynamics by NMR Spectroscopy
RNA is a flexible molecule: This is what makes it functional, but also difficult to study, especially when the goal is to access those dynamic, often low-populated and short-lived conformers. Here th...
One misconception about RNA is that it’s static. RNA is actually ✨dynamic✨ and exists as a ground state in exchange with higher energy excited states. These #RNAdynamics allow RNA to perform functions and can be studied at the atomic level using NMR. Read more here 👉 bit.ly/43kbBQ3 #scicommUU25
20.05.2025 15:47 — 👍 2 🔁 0 💬 0 📌 0
Hi BSKY!💫 I'm Katja, a 2nd year PhD student at Uppsala University in the Petzold group. My projects focus on improving #in-cellNMR of RNA & using #solid-stateNMR to investigate RNA:protein interactions #scicommUU25
20.05.2025 15:45 — 👍 5 🔁 0 💬 0 📌 0



