PNAS
Proceedings of the National Academy of Sciences (PNAS), a peer reviewed journal of the National Academy of Sciences (NAS) - an authoritative source of high-impact, original research that broadly spans...
Congratulations to MD-PhD student, Amanda Velez and all co-authors on our new PNAS paper describing how the innate immune protein calprotectin incapacitates autolysins, inducing tolerance to B-lactam antibiotics. A fun collaboration with Thomas Kehl-Fie. www.pnas.org/doi/10.1073/...
14.01.2026 17:24 — 👍 17 🔁 4 💬 0 📌 0

Schematic overview of the process of DNA-affinity / pull down identification of unknown nucleic acid-binding proteins
Identification of Previously Unknown DNA-Binding Proteins Using DNA Affinity/Pull-Down Methods Followed by Mass Spectrometry
Jutras, Babb, Jusufovic, Krusenstjerna, Saylor, Verma, and Stevenson
Current Protocols 2025, 5:e70264
doi: 10.1002/cpz1.70264
#MicroSky
26.11.2025 16:00 — 👍 39 🔁 10 💬 1 📌 1

🚨Preprint alert - this is a big one! We transfer the revolutionary power of TnSeq to bacteriophages.
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8
20.11.2025 20:39 — 👍 210 🔁 90 💬 11 📌 5
Congratulations Alastair and Prerna! Exciting times :)
12.11.2025 12:21 — 👍 1 🔁 0 💬 1 📌 0
Congratulations, Jay! Looking forward to learning about your discoveries :)
31.10.2025 08:09 — 👍 1 🔁 0 💬 0 📌 0
An Rhs effector uses distinct target cell functions to intoxicate bacterial and fungal competitors https://www.biorxiv.org/content/10.1101/2025.10.28.685041v1
29.10.2025 02:18 — 👍 3 🔁 2 💬 0 📌 0
Independent research fellowships leading to tenured positions at the John Innes Centre.
Repost = nice. Thank you very much!!!
03.10.2025 16:06 — 👍 49 🔁 87 💬 0 📌 4
Cool!
29.09.2025 23:10 — 👍 0 🔁 0 💬 0 📌 0
Congrats, Lev!
07.06.2025 19:35 — 👍 1 🔁 0 💬 1 📌 0
They will survive much better if you let them grow into stationary phase in rich media first, and then transfer them to a minimal medium with no carbon source. Transferring to carbon starvation directly from exponential growth causes faster death. See for example PMID: 32500952 and PMID: 34058747.
23.04.2025 20:41 — 👍 2 🔁 0 💬 0 📌 0
yes, the MMBs studied by george schaible and roland hatzenpichler are fascinating 𝗼𝗯𝗹𝗶𝗴𝗮𝘁𝗲 𝗺𝘂𝗹𝘁𝗶𝗰𝗲𝗹𝗹𝘂𝗹𝗮𝗿 bacteria 🤩
STC had mentioned them first in 2007 > schaechter.asmblog.org/schaechter/2...
and recently in > schaechter.asmblog.org/schaechter/2...
#MicroSky
09.04.2025 13:04 — 👍 15 🔁 6 💬 1 📌 1
Future Directions of the Prokaryotic Chromosome Field
In September 2023, the Biology and Physics of Prokaryotic Chromosomes meeting ran at the Lorentz Center in Leiden, The Netherlands. As part of the workshop, those in attendance developed a series of ....
In @molecularmicro.bsky.social: two special issues on Prokaryotic Chromosome Organization. Many excellent contributions, including a vision article: 'Future directions of the Prokaryotic Chromosome Field', outcome of BioPhyChrom 2023 @biophychrom.bsky.social onlinelibrary.wiley.com/doi/10.1111/...
20.02.2025 20:14 — 👍 32 🔁 25 💬 2 📌 2
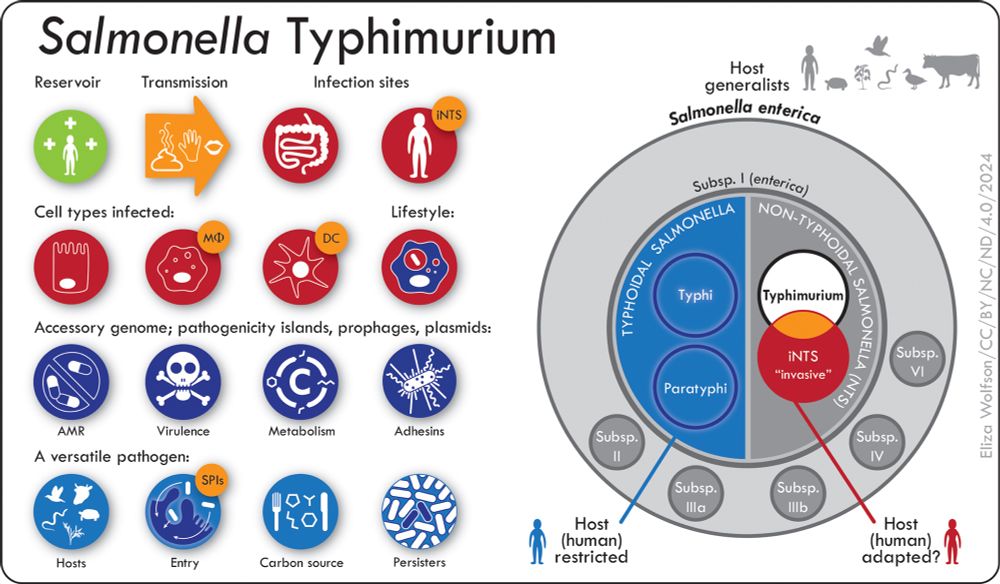
The taxonomy & lifestyle of Salmonella Typhimurium
www.microbiologyresearch.org/content/jour...
23.01.2025 22:56 — 👍 22 🔁 9 💬 0 📌 0

Congrats to Findlay and many thanks to examiners Dan Neill and Martin Welch for their work on a successful viva today! Findlay has worked so hard and accomplished so much while blazing the trail as the first PhD student in the lab :)
16.01.2025 17:22 — 👍 2 🔁 0 💬 0 📌 0
Together with @frimanscience.bsky.social and our Chinese colleagues, we have worked out how to predict
(i) siderophore molecular structures
www.biorxiv.org/content/10.1...
and
(ii) siderophore interaction networks in communities
www.biorxiv.org/content/10.1...
from sequence data!
09.11.2023 12:13 — 👍 25 🔁 18 💬 2 📌 1
Thanks for the introduction!
08.11.2023 23:27 — 👍 1 🔁 0 💬 0 📌 0
microbiologist, McMaster U 🇨🇦 • type IV pili, phages, biofilms, antibiotic resistance • equestrian🐴 • cancer survivor🎗️
AMR researcher, pharmacodynamicist and broad interest in maintaining antibiotics as critical societal assets.
Bringing together the best of research and industry to overcome one of the biggest threats facing humanity.
We (The World Health Organization) are the United Nations’ health agency championing Health For All. Always check the latest posts for updated advice/information. We will remove misinformation, spam, and hate speech here.
TARGetAMR is a UK-based research network funded by UK Research and Innovation (UKRI) with the primary goal of using genomics to tackle antimicrobial resistance (AMR).
Associate Professor at Utrecht University, Infectious Diseases & Immunology. (meta)genomes, AMR, zoonotic pathogens. Cookie and cake connoisseur.
publications: https://scholar.google.com/citations?hl=nl&user=VHHhpoAAAAAJ&view_op=list_works&sortby=pubdate
Professor of Molecular Diagnostics and Infection at University of Edinburgh | AMR Strategy Lead Edinburgh Infections Diseases | Coordinator DOSA Project | Co-Lead Translational Medicine | working with great teams
Bacteria lover 🦠🧫🔬
PhD. in Molecular Microbiology 🇬🇧 Biochemist 🇨🇱
Institute of Public Health Chile 🇨🇱
RNA modifications, antibiotic tolerance, bacterial stress, non coding RNAs, Vibrio cholerae.
Institut Pasteur & EGM (IBPC, CNRS)
Principal infectious disease modeller at UKHSA, specialising in transmission modelling and computational immunology. Researching AMR and HCAI. Ironman triathlete, runner, cyclist, cold water swimmer, mum to a toddler, and animal lover
CEO, UKHSA
Infectious diseases clinician and epidemiologist
Hon Consultant Infectious Diseases, Royal Free London
Hon Prof Infectious Diseases & Health Security, UCL
Contains multitudes. Global health lawyer and advisor. PhD candidate at LSHTM researching the global politics of the use of antibiotics. 🌍💊 Consultant at WHO + FAO + WOAH, ex-MSF
Infectious Diseases physician | Hospital Epidemiologist | Denver Health (Hospital) and University of Colorado | USA ESCMID Local Champion | Passionate about diagnostic stewardship, molecular epi, and AI
Clinical pharmacist interested in infectious disease, antimicrobial resistance, and optimizing PK/PD targets
Also an avid birder and photographer
María de Toro. Bioinformatician. Sequencing Microbial genomes 🧬. Manager of a Genomics & Bioinformatics Core Facility
#NoSciNoFuture #OneHealth🌏 #FoodSafety #Bioinformatics #MGEs #plasmids
❗️Only science
Professor for Mucosal Immunology, ETH Zürich, Switzerland, and Sir William Dunn School of Pathology, Oxford, UK. We work on novel oral vaccines, AMR, and microbiome function. Making science fairer, kinder and more equitable 🏳️🌈🏳️⚧️. Views are my own.
Professor of infectious disease epidemiology at Centre for Global Health Research, Oxford. 🤓🧀☕️🚴🏼🦠💉💊
Animal scientist PhD ABD. Vet micro Gram -ve bacteria, AMR, One Health, Zoonoses & Bioinformatics. MEd student. Head tutor. Multiply Disabled, cPTSD, AuDHD. Animal lover. Spoonie. Enby. Pan. Ace. Always carry dog treats.
https://linktr.ee/DavidWakeham
Lead for Molecular Microbiology @sheffielduni.bsky.social
@molmicrosheffield.bsky.social
#SEN parent, #AngelmanSyndrome
Clostridial cell biology, AMR & bacteriophage structure and function
He/Him
https://sites.google.com/sheffield.ac.uk/fagan-lab/








