
"Bulk RNA-seq to the rescue of differential expression analysis in single-cell transcriptomics" 🧫 📑 A new PCI Genomics recommendation by @mireyaplass.bsky.social: doi.org/10.24072/pci...
01.12.2025 08:30 — 👍 2 🔁 5 💬 1 📌 0@markrobinsonca.bsky.social
statistical bioinformatics; canadian / swiss; #methodsmatter #rstats; open science advocate; grumpy towards prestige worshippers, excessive admin and bros; will sacrifice sleep for a sunrise. https://robinsonlabuzh.github.io/

"Bulk RNA-seq to the rescue of differential expression analysis in single-cell transcriptomics" 🧫 📑 A new PCI Genomics recommendation by @mireyaplass.bsky.social: doi.org/10.24072/pci...
01.12.2025 08:30 — 👍 2 🔁 5 💬 1 📌 0

Orchestrating Spatial Transcriptomics Analysis with Bioconductor
(including interoperability with Python)
Paper: www.biorxiv.org/content/10.1...
Book: bioconductor.org/books/OSTA/
#Rstats
Many people hours, calls and messages later: OSTA is now “in (pre)print”, though the real thing lives at bioconductor.org/books/OSTA.
Check it out, get in touch. We welcome any feedback, suggestions, wishes (& contributions).
It’s been a joy working with you @estellayixingdong.bsky.social!
We are so excited to announce that our latest paper is out in @jem.org
By combining single-cell RNA sequencing and 3D imaging, we reveal striking structural and molecular differences between human and mouse dermal lymphatic vasculature, opening up new perspectives on #Lymphatics and #HumanBiology

From @csoneson.bsky.social , @markrobinsonca.bsky.social & colleagues in @plos.org #Computational #Biology | Building a continuous #benchmarking ecosystem in #bioinformatics | #Education 🧬 🖥️ 🧪 🔓
⬇️
journals.plos.org/ploscompbiol...
Hybrid untargeted and targeted RNA sequencing facilitates genotype-phenotype associations at single-cell resolution #SingleCell 🧪🧬🖥️
https://www.biorxiv.org/content/10.1101/2025.11.06.686971v1

DESpace2: detection of differential spatial patterns in spatial omics data. #SpatialOmics #DifferentialSpatialPatterns #scRNAseq #Genomics #Bioinformatics @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...

🧬 EuroBioC2026 is coming to Turku, Finland, from June 3–5, 2026!
Expect 3 exciting days with the Bioconductor community, showcasing the latest advances in Bioconductor software and emerging technologies shaping computational biology.
👉 More details soon at eurobioc2026.bioconductor.org
.. where some compelling demonstration of performance/usefulness is made (ideally done in a way that is interoperable with community efforts) .. and over the longer term, communities complement/consolidate with continuous benchmarks. In principle, Omnibenchmark supports both use cases.
24.10.2025 14:38 — 👍 1 🔁 0 💬 1 📌 0Interesting discussion! I agree that "extensive" benchmarks within method papers are not really the priority/essential. But at the same time, it would be tough to motivate having benchmark-less methods papers. I think we need a middle ground ..
24.10.2025 14:38 — 👍 1 🔁 0 💬 1 📌 0I'm very excited to share our latest preprint!
We introduce structure-based analysis of spatial omics data – an approach that focuses on multi-cellular anatomical structures rather than single cells.
We also present sosta to facilitate this type of analysis: bioconductor.org/packages/sos...
Analysis of anatomical multi-cellular structures from spatial omics data using sosta https://www.biorxiv.org/content/10.1101/2025.10.13.682065v1
14.10.2025 14:48 — 👍 1 🔁 1 💬 0 📌 1
Looking for scientists working with long-read transcriptomics technologies to join a COST action proposal. Contact us!!! @nanoporetech.com @pacbio.bsky.social
02.10.2025 17:24 — 👍 6 🔁 6 💬 0 📌 1There are some similarities, but also some differences.
Scale: cells versus regions, but probably more importantly, Feature Space: segmentation focuses a lot on DNA and cell membrane markers (possibly transcript locations), while SAC is more about multi-cellular / neighbourhoods.

Martin Emons on SpatialFDA - searching for differential colocalization in spatial omics data
18.09.2025 09:46 — 👍 9 🔁 1 💬 0 📌 0
EuroBioC2025 group photo on PRBB terrace with sea in background.

EuroBioC2025 welcome in PRBB auditorium by host Robert Castelo, showing slides with conference sticker and sponsor logos.

EuroBioC2025 hexwall banner with the hexwall creator Kevin Rue-Albrecht standing beside it.

EuroBioC2025 carved wooden keyring, a gift for all participants, created by Annekathrin Nedwed.
EuroBioC2025 is in full swing here in Barcelona!
Day 1 was full of great talks, conversations, and that amazing Bioconductor community energy. Excited for what’s ahead today and tomorrow!
#EuroBioC2025 #Bioconductor #OpenScience #Community

We are excited to share the publication of our paper on exploratory spatial statistics for spatial omics data
academic.oup.com/nar/article/...
MIMIC: a flexible pipeline to register and summarize IMC-MSI experiments https://www.biorxiv.org/content/10.1101/2025.07.08.663623v1
12.07.2025 05:47 — 👍 3 🔁 1 💬 0 📌 0
Update: We greatly revised our paper and renamed it “Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta”.
We discuss the broad range of exploratory spatial statistics options for spatial Omics technologies and show relevant use cases.
arxiv.org/abs/2412.01561

Did you ever notice discrepancies in benchmarking of bioinformatics tools? We did too! Setting out to benchmark spatial clustering methods, we encountered major reproducibility issues in previous benchmarks and questionable "ground truths".
More in our preprint: www.biorxiv.org/content/10.1...
DESpace2: detection of differential spatial patterns in spatial omics data https://www.biorxiv.org/content/10.1101/2025.06.30.662268v1
02.07.2025 17:46 — 👍 6 🔁 3 💬 0 📌 0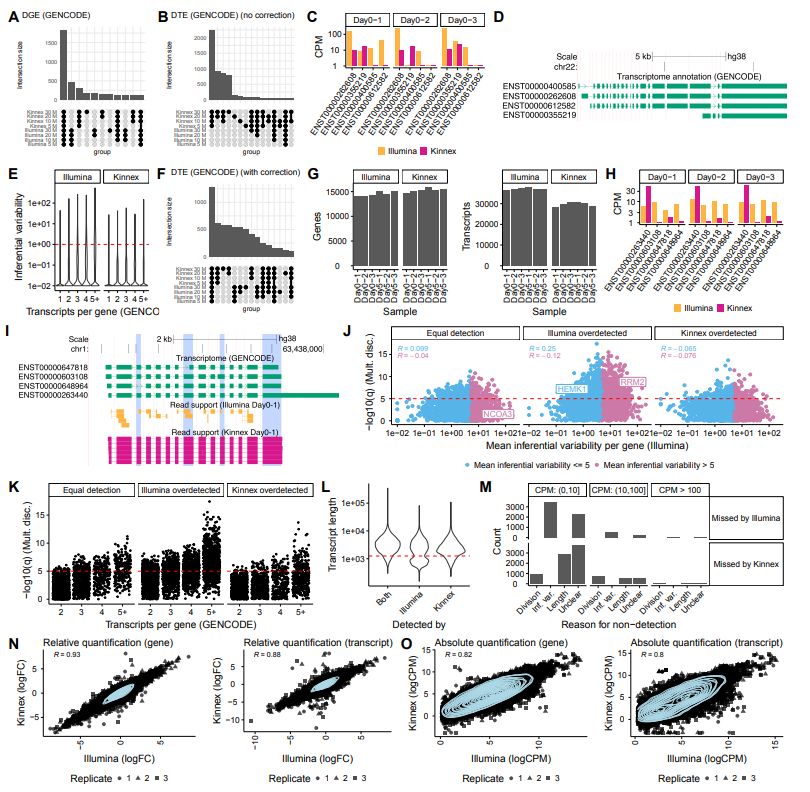
A Systematic Benchmark of High-Accuracy PacBio Long-Read RNA Sequencing for Transcript-Level Quantification. #HiFi #LongReads #Sequencing #Transcriptomics
@pacbio.bsky.social @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
🤖 SIB supplies the foundation of transformational AI by providing:
⭐ gold-standard training data through curated databases
🔗 connected information through knowledge representation
💪 trustworthy evaluation through benchmarking.
See how in the #SIBProfile 2025: issuu.com/sibswissinst...
![[2025-06-11] Pseudobulk DGE statistical methods journal club](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:h7p2smbyfobkpcs7dbgvhmeh/bafkreifypyga25y52cts32z2bfhpgmx4znvlfjafg2b2vsvcnlw3hs4qvy@jpeg)
Pseudobulk DGE #RStats journal club
* #muscat doi.org/10.1038/s414... @helucro.bsky.social
* pbDD doi.org/10.1101/2023... @davi1893.bsky.social
* #dreamlet doi.org/10.1101/2023... @panosroussos.bsky.social
* #distinct doi.org/10.1214/22-A... @markrobinsonca.bsky.social
youtu.be/TB9dfKPfns8
Guest Editors:
Mark Robinson, University of Zurich
Fritz Joachim Sedlazeck, Baylor College of Medicine
Hong-Bin Shen, Shanghai Jiao Tong University
Jean Yee Hwa Yang, The University of Sydney
Xin Maizie Zhou, Vanderbilt University
Heads-up that Genome Biology is starting a new benchmark collection ("Benchmarks v2.0"):
www.biomedcentral.com/collections/...

@jeffmold.bsky.social @willmacnair.bsky.social would be very interesting indeed. Here is the classic study doing something along these lines (not in genomics):
journals.sagepub.com/doi/10.1177/...

Honored to deliver the opening keynote talk at the Modern Benchmarking Conferene in Ascona a very much needed meeting organizad by @markrobinsonca.bsky.social @iscb.bsky.social sites.google.com/view/ascona2...
24.03.2025 09:07 — 👍 5 🔁 3 💬 0 📌 0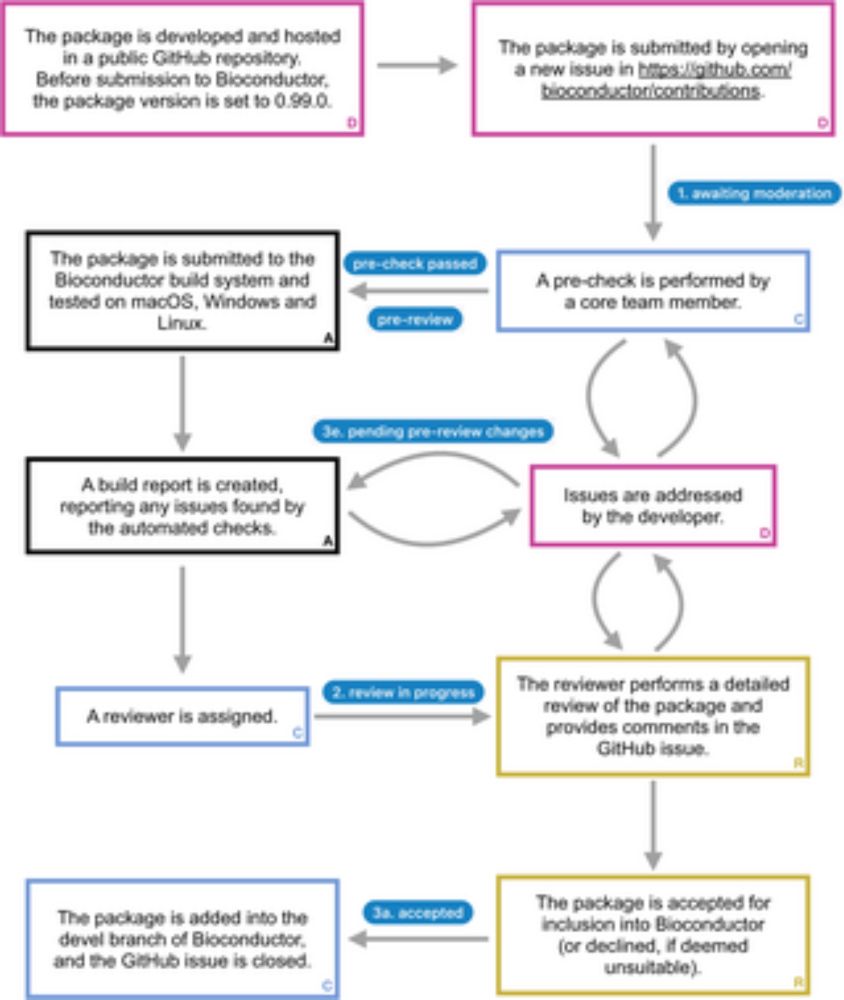
From @csoneson.bsky.social , @mramos148.bsky.social, @jorainer.bsky.social & colleagues in @plos.org #Computational #Biology | Eleven quick tips for writing a #Bioconductor package | #Bioinformatics #Education #Rstats #PLOSCBQT 🧬 🖥️ 🧪 🔓
⬇️
journals.plos.org/ploscompbiol...
Quick update. We have a pretty exciting lineup of topics/speakers in and around benchmarking that will be presented in Ascona at the end of the month:
sites.google.com/view/ascona2...
There are few registration slots left, so if you are interested to join us, get in touch.