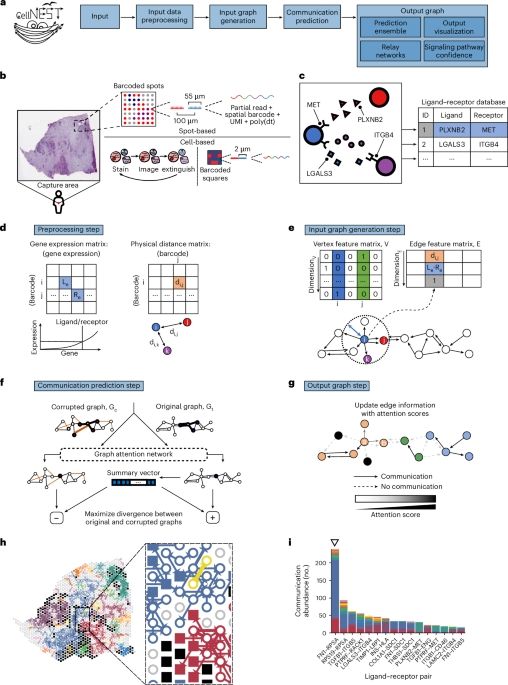
CellNEST reveals cell–cell relay networks using attention mechanisms on spatial transcriptomics👍
www.nature.com/articles/s41...
@lukasvalihrach.bsky.social
Single-Cell & Spatial Transcriptomics | Group Leader at GliaOmics Lab | Head of GeneCore Facility | Co-founder at Carta Genum | Glial Cells in Health & Disease

CellNEST reveals cell–cell relay networks using attention mechanisms on spatial transcriptomics👍
www.nature.com/articles/s41...
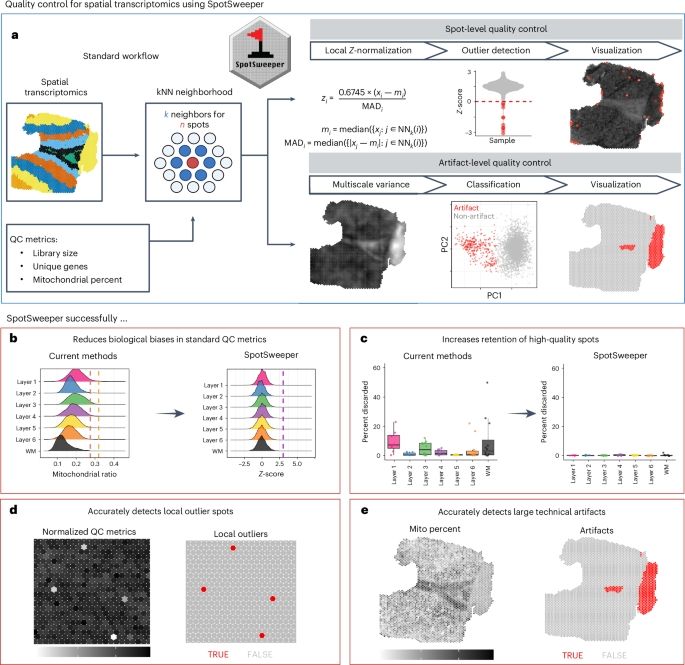
SpotSweeper: spatially aware quality control for spatial transcriptomics👍
www.nature.com/articles/s41...
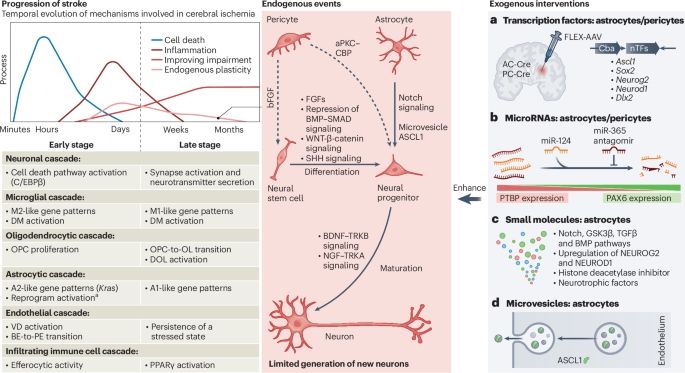
Changing genes, cells and networks to reprogram the brain after stroke🫶
www.nature.com/articles/s41...

KODAMA enables self-guided weakly supervised learning in spatial transcriptomics
www.biorxiv.org/content/10.1...
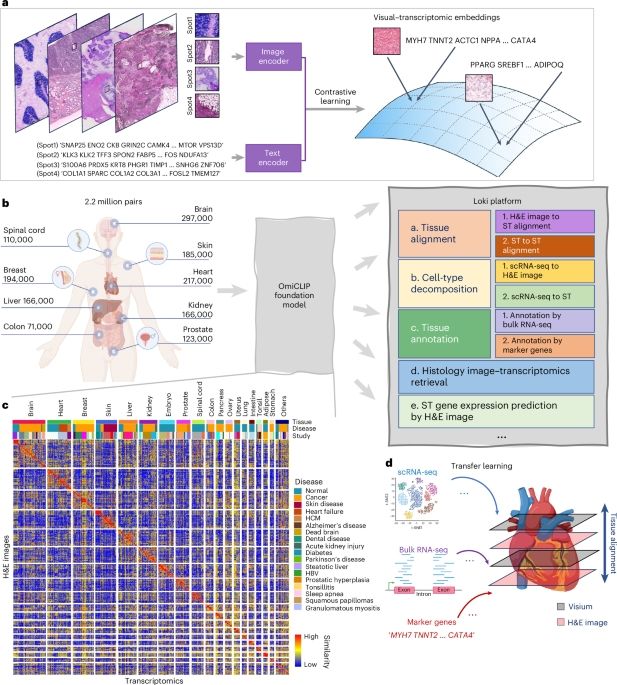
A visual–omics foundation model to bridge histopathology with spatial transcriptomics🔥🔥🔥
www.nature.com/articles/s41...

AI-driven 3D Spatial Transcriptomics🤯
arxiv.org/abs/2502.17761
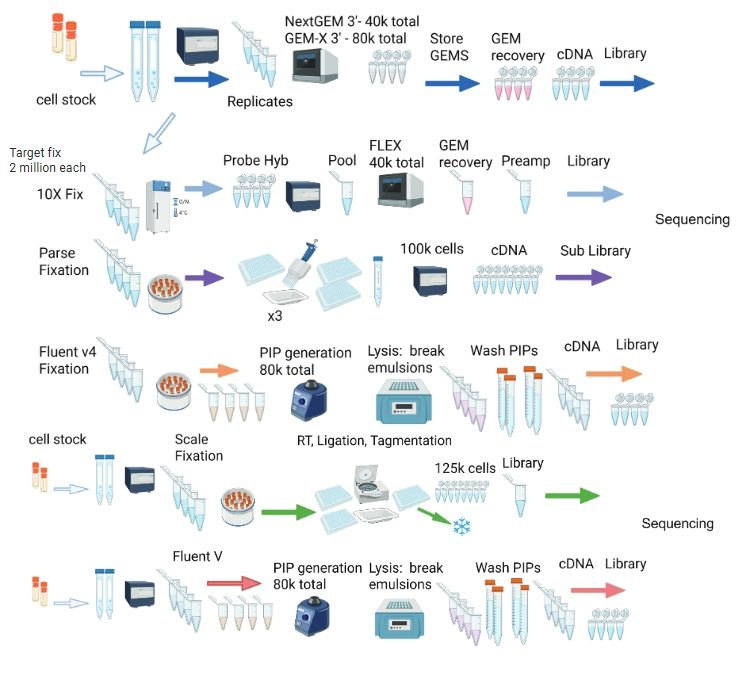
Evaluating the practical aspects and performance of commercial single-cell RNA sequencing technologies🔥
www.biorxiv.org/content/10.1...
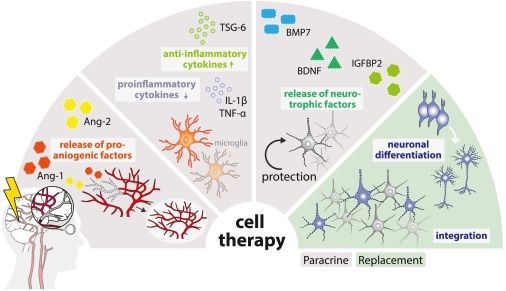
How neural stem cell therapy promotes brain repair after stroke🫶
www.cell.com/stem-cell-re...
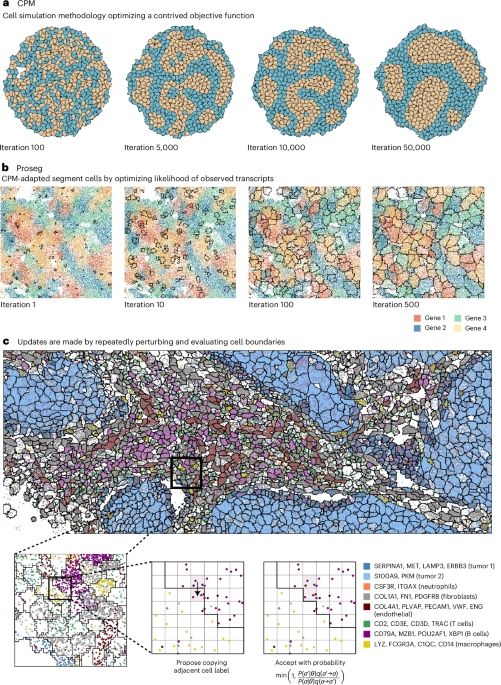
Cell simulation as cell segmentation⚡
www.nature.com/articles/s41...
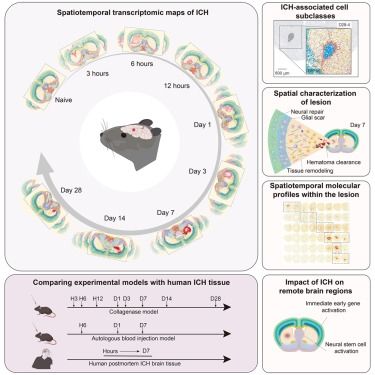
Spatiotemporal transcriptomic maps of mouse intracerebral hemorrhage at single-cell resolution🔥
www.cell.com/neuron/abstr...
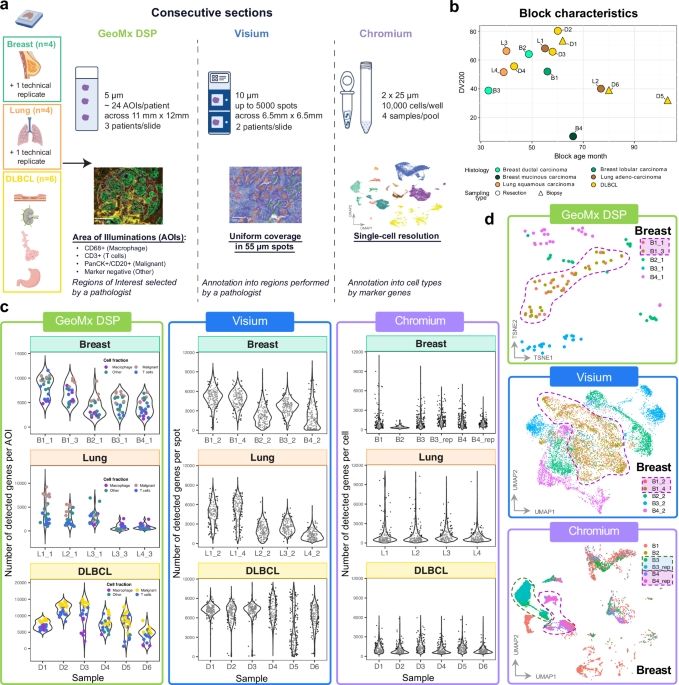
Transcriptome analysis of archived tumors by Visium, GeoMx DSP, and Chromium reveals patient heterogeneity🙌
www.nature.com/articles/s41...
Beyond cell atlases: spatial biology reveals mechanisms behind disease🫶
www.nature.com/articles/s41...
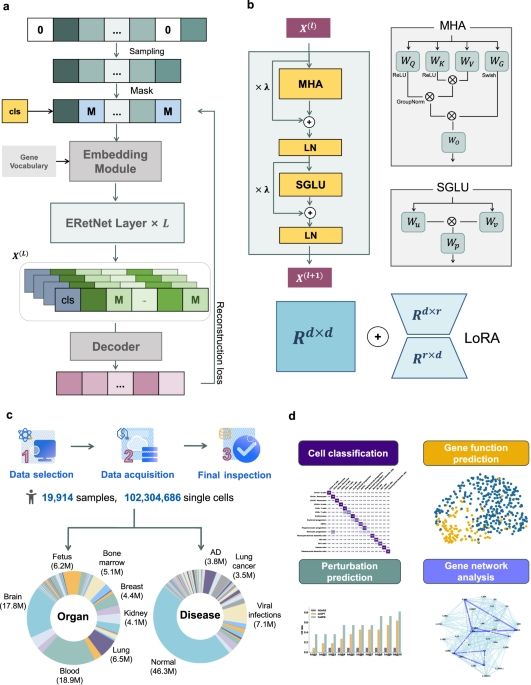
CellFM: a large-scale foundation model pre-trained on transcriptomics of 100 million human cells🤯
www.nature.com/articles/s41...
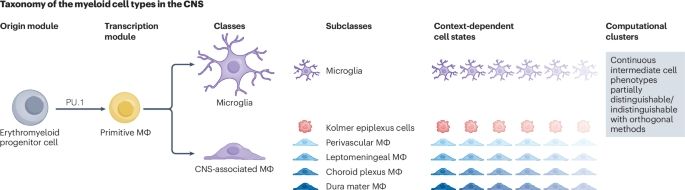
A dynamic and multimodal framework to define microglial states🔥
www.nature.com/articles/s41...
Cautions on utilizing plasma GFAP level as a biomarker for reactive astrocytes in neurodegenerative diseases
molecularneurodegeneration.biomedcentral.com/articles/10....

A spatiotemporal cancer cell trajectory underlies glioblastoma heterogeneity🔥
www.biorxiv.org/content/10.1...
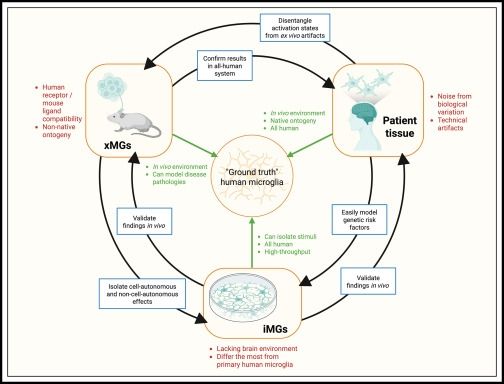
Microglia transcriptional states and their functional significance: Context drives diversity🫶
www.cell.com/immunity/ful...
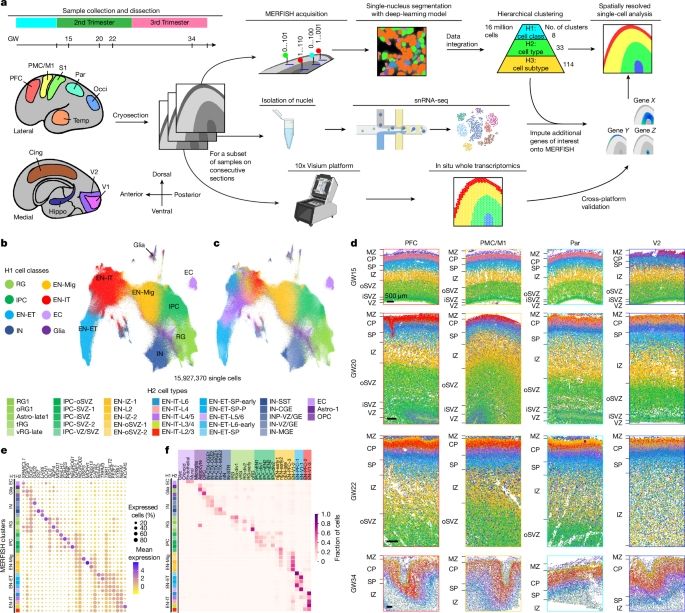
Spatial transcriptomics reveals human cortical layer and area specification🧠
www.nature.com/articles/s41...
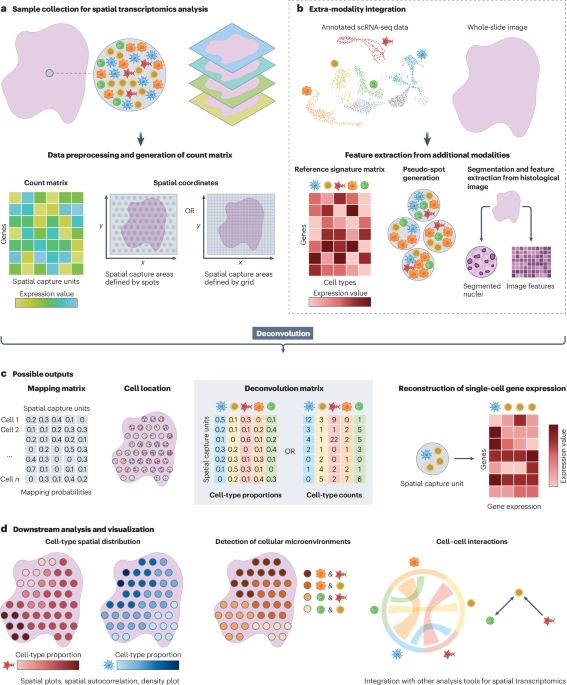
Cell-type deconvolution methods for spatial transcriptomics🔥
www.nature.com/articles/s41...
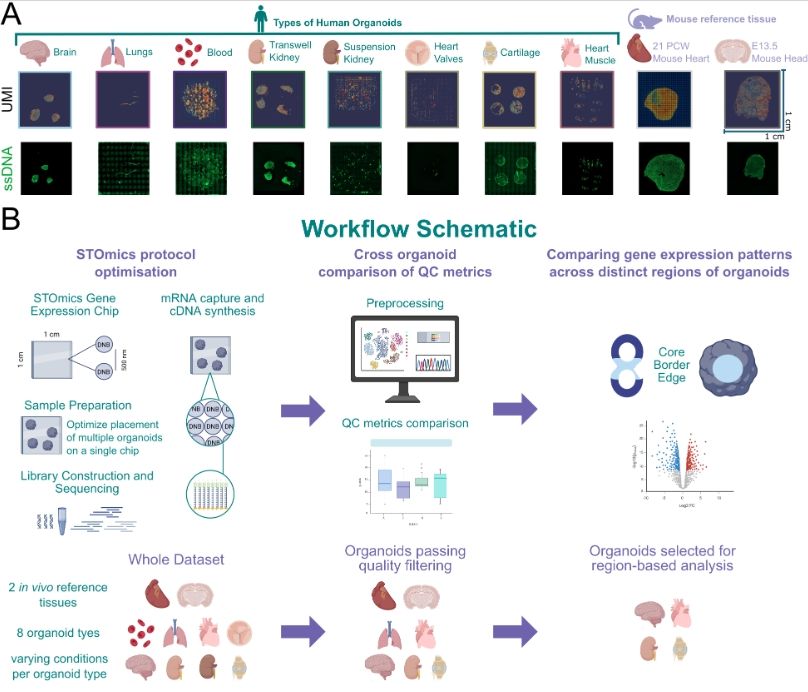
Application of spatial transcriptomics across organoids: a high-resolution spatial whole-transcriptome benchmarking dataset
www.biorxiv.org/content/10.1...

EnrichSci: Transcript‑guided Targeted Cell Enrichment for Scalable Single‑Cell RNA Sequencing
www.biorxiv.org/content/10.1...
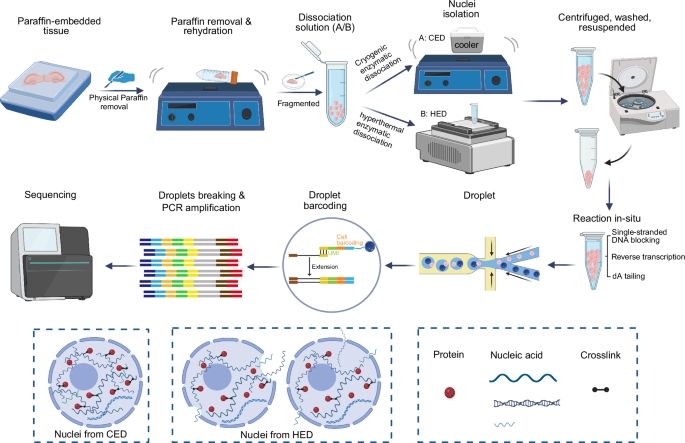
snCED-seq: high-fidelity cryogenic enzymatic dissociation of nuclei for single-nucleus RNA-seq of FFPE tissues
www.nature.com/articles/s41...

Next generation statistical framework for next generation spatial transcriptomics data
www.biorxiv.org/content/10.1...

A computational framework for mapping isoform landscape and regulatory mechanisms from spatial transcriptomics data 🤔
www.biorxiv.org/content/10.1...

ShinyCell2: An extended library for simple and sharable visualisation of spatial, peak-based and multi-omic single-cell data
www.biorxiv.org/content/10.1...
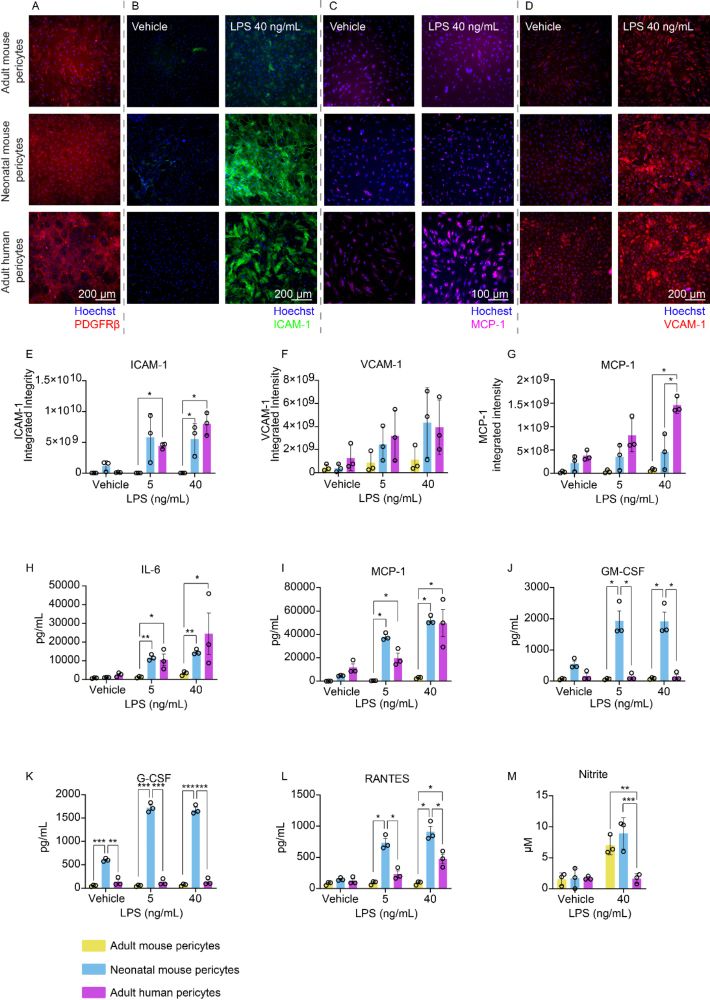
Heterogeneity in pericyte inflammatory responses across age and species highlight the importance of human cell models
molecularbrain.biomedcentral.com/articles/10....
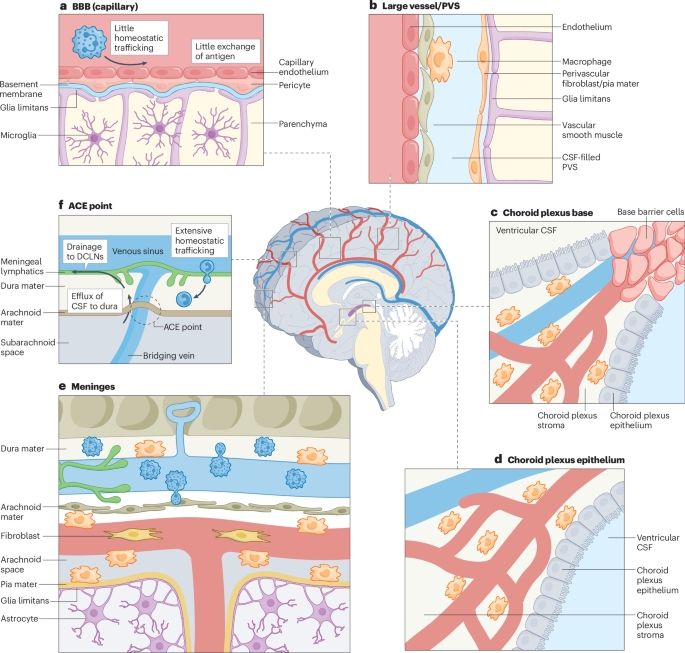
Redefining CNS immune privilege🫶
www.nature.com/articles/s41...

How the Human Cell Atlas is fast-tracking new medicines😍
www.npr.org/2025/04/18/g...

SpatialAgent: An autonomous AI agent for spatial biology🤯
www.biorxiv.org/content/10.1...
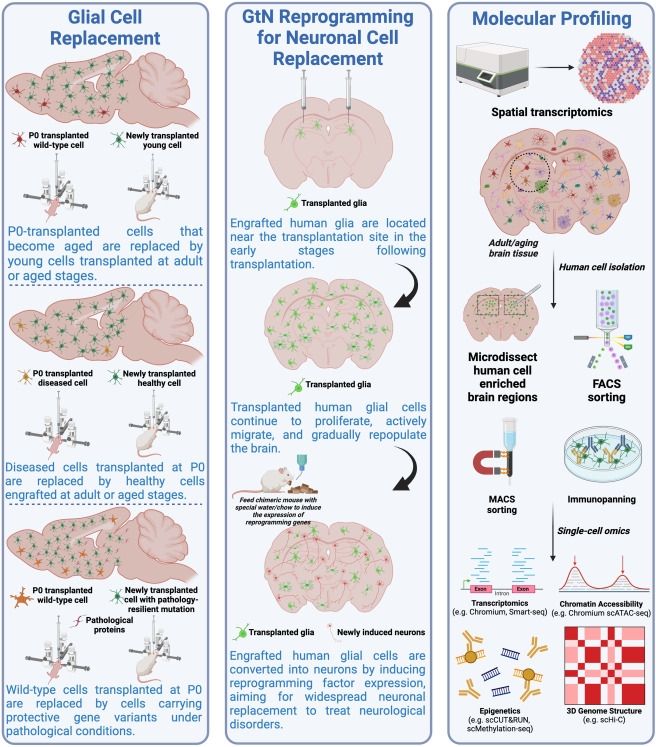
Chimeric brain models: Unlocking insights into human neural development, aging, diseases, and cell therapies
www.cell.com/neuron/abstr...