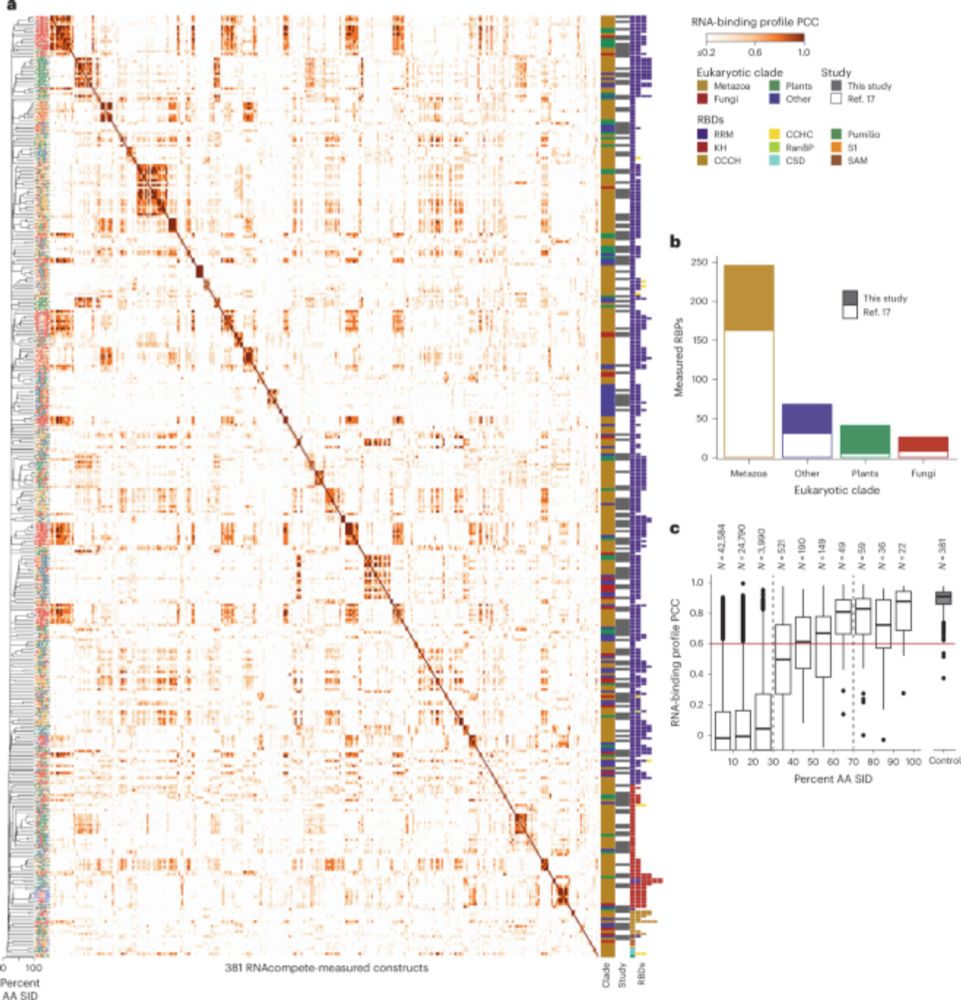
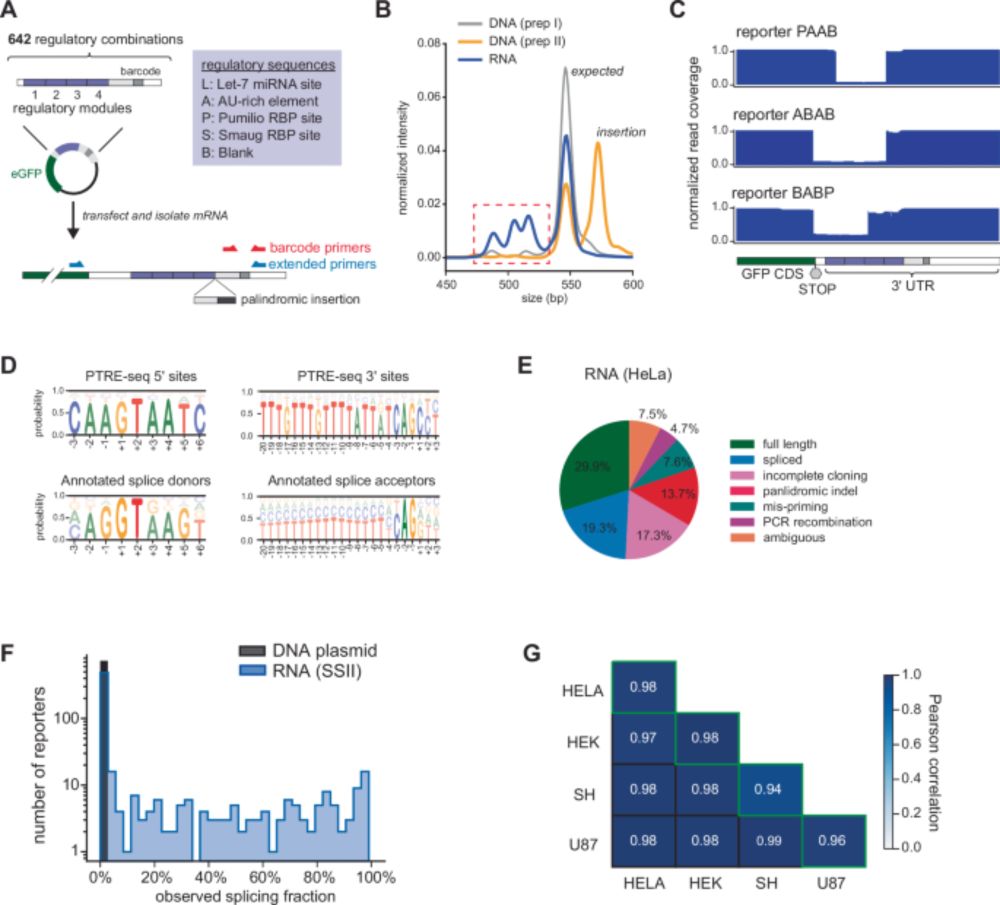
Anoctamin-2-specific T cells link Epstein-Barr virus to multiple sclerosis
Researchers identified anoctamin-2 (ANO2) as a frequent autoimmune target in multiple
sclerosis, with T cell responses against ANO2 occurring in over half of patients.
These ANO2-specific T cells shar...
Epstein-Barr Virus #EBV was linked to #MultipleSclerosis - now a plausible cause has been found, misidentification by longterm memory T-cells that pick on the wrong protein, ANO2, instead of the EBV-antigen. Massive inflection towards the elimination of MS!!!
🧪🧠Cell www.cell.com/cell/fulltex...
15.01.2026 01:41 — 👍 872 🔁 283 💬 12 📌 49

In a new work with Joseph Rich and Conrad Oakes we tackle the problem of how to best organize alluvial plots. We formalize two optimization problems and develop a solution for them based on the neighbornet algorithm, implemented in the program wompwomp: github.com/pachterlab/w...
05.09.2025 12:20 — 👍 33 🔁 10 💬 3 📌 0

🚀 Two exciting days of Computational Biology at the Cardio-Pulmonary Institute in Bad Nauheim @cpi-exstra.bsky.social ! 🔬 Workshop on single-cell analysis + the CPI repository data ressource; 💡 Hackathon together with the Schulz Lab @themarcelschulz.bsky.social
Collaboration, coding & discovery
28.08.2025 12:07 — 👍 6 🔁 3 💬 0 📌 0

🎉 To mark the fourth extension of the CPI, we celebrated our success at the @mpi-hlr.bsky.social 🥳 We are delighted about the renewed funding and would like to thank everyone who made this step possible! Read more in the latest issue of "Synapse": www.unimedizin-ffm.de/fileadmin/re...
04.08.2025 06:57 — 👍 6 🔁 6 💬 1 📌 0
we asked a simple question:
What does it take to learn the rules of RNA base pairing?
using standard deep-learning technics, got a simple answer:
don't need structures, nor alignments or many parameters
only a few RNA sequences and 21 parameters;
doi.org/10.1101/2025...
02.08.2025 20:39 — 👍 20 🔁 9 💬 2 📌 1
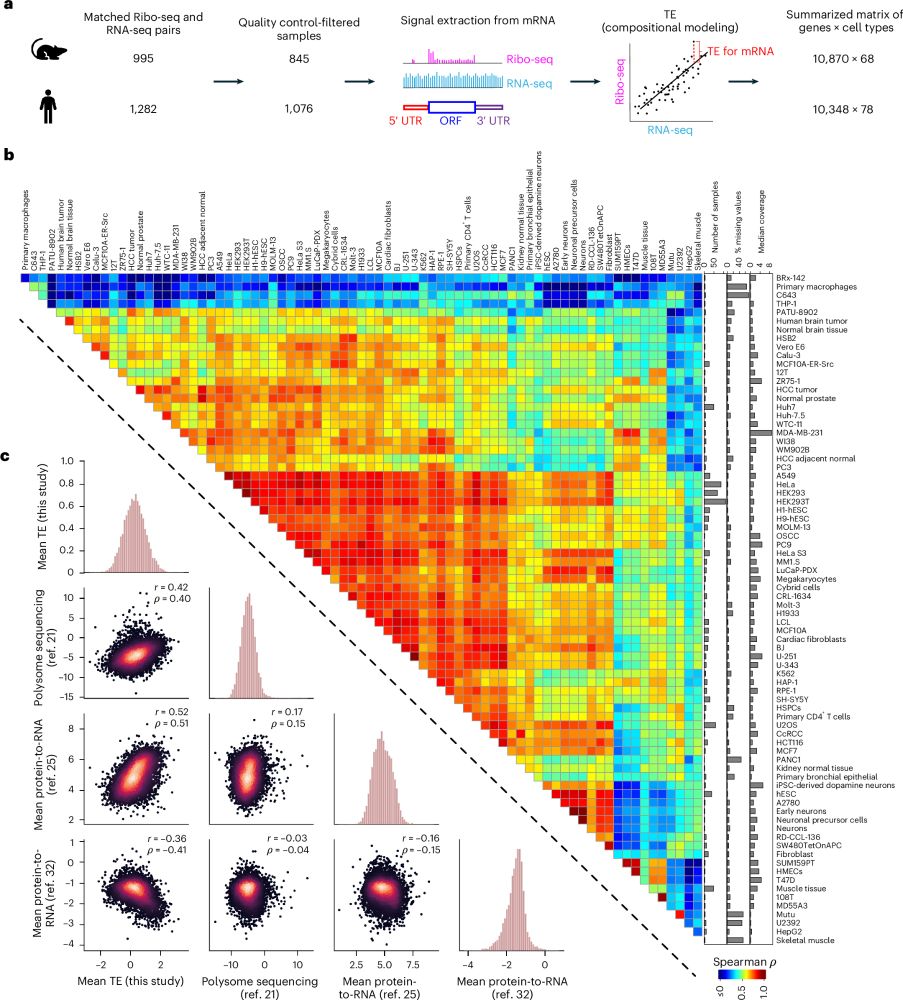
Predicting the translation efficiency of messenger RNA in mammalian cells
Nature Biotechnology - A deep convolutional neural network model predicts the influence of the full-length mRNA sequence on translation efficiency.
Very excited that our most significant work, a collaboration w/ Dr. Can Cenik at UT Austin on translational gene regulation, was finally published in Nature Biotechnology in a dual set of studies:
Paper 1 -- an AI model trained to predict translation rates from mRNA sequences: rdcu.be/exN1l
25.07.2025 13:37 — 👍 49 🔁 21 💬 3 📌 0
And BIG thanks to #RegSys track organizers @abardet.bsky.social A. Emad J. Welch R. Gordan @xiuweizhang.bsky.social R. Beekman @martamele.bsky.social @anniquec.bsky.social @shaunmahony.bsky.social @ferhatay.bsky.social and co-chairs @alemedinarivera.bsky.social and @themarcelschulz.bsky.social
24.07.2025 19:00 — 👍 7 🔁 1 💬 0 📌 0
What a fantastic #ISMBECCB2025 conference it has been, with an outstanding #RegSys @iscb-regsys.bsky.social COSI track!
Thanks, everyone, for submitting, attending, presenting, and organizing @iscb.bsky.social @eccb-europe.bsky.social
24.07.2025 19:00 — 👍 9 🔁 4 💬 2 📌 0

🧬 New @emblebi.bsky.social service beta launched:
Perturbation Catalogue
🔎 Browse genetic perturbation datasets
🧠 Train models
🧬 Interpret variant function
🧭 Explore gene dependencies
Search, test APIs, suggest datasets, send feedback!
🔗 www.ebi.ac.uk/perturbation...
Funded by @opentargets.org
23.07.2025 10:06 — 👍 44 🔁 18 💬 1 📌 0

Last talk at #regsys today is a keynote by Mafalda Dias about modelling sequence variation in Evolution with insights into diseases #ismbeccb2025
23.07.2025 16:23 — 👍 4 🔁 0 💬 0 📌 0

First talk of the last session of RegSys day 1 at #ismb2025 is from Jacob Schreiber on how we read the book of DNA
Spoiler: regulation is key
23.07.2025 15:43 — 👍 3 🔁 1 💬 0 📌 1

Laura Gunsalus talks about design of small native regulatory elements using #ML #ismbeccb2015 #regsys
23.07.2025 16:03 — 👍 0 🔁 0 💬 0 📌 0

Last talk in this #regsys session is Jishnu Das who talks about regulatory programs that determine B-cell fates #ismbeccb2025
23.07.2025 14:43 — 👍 1 🔁 0 💬 0 📌 0

Tatevik Jalatyan will talk about a new pipeline for estimating cell cycle phase from single cell data #ismbeccb2025 #regsys
23.07.2025 14:24 — 👍 0 🔁 0 💬 0 📌 0

Surag Nair talks about a general framework to combine different paradigms of DL genome models #ismbeccb2025 #regsys
23.07.2025 14:03 — 👍 0 🔁 0 💬 0 📌 0

Vitalii Kleshchenikow about a biophysical model for learning gene regulation from single cell data #ismbeccb2025 #regsys @oliverstegle.bsky.social
23.07.2025 13:45 — 👍 0 🔁 0 💬 0 📌 0
#regsys session is overflowing at #ismbeccb2025 consider to go to rooms 6 and 7 for video broadcast if full
23.07.2025 13:40 — 👍 1 🔁 1 💬 0 📌 0

Laura Rumpf talks about the MetaFR pipeline to learn gene regulation models from unpaired and paired scATAC and scRNA data #ismbeccb2025 #regsys #schulzlab @dzhk.de @sfb1531.bsky.social
23.07.2025 13:21 — 👍 3 🔁 2 💬 0 📌 0
Refreshed after lunch we start with Oluwatosin Oluwadare to introduce a new approach for single cell HiC analysis #ismbeccb2025 #regsys
23.07.2025 13:04 — 👍 1 🔁 1 💬 0 📌 0

Last speaker before lunch is Christophe Vroland to talk about confounders in Rna-Dna interaction data analysis #regsys #ismbeccb2025
23.07.2025 12:00 — 👍 2 🔁 0 💬 0 📌 0

Laura Hinojosa talks about TFs that regulate replication timing
#RegSys #ismbeccb2025 @ferhatay.bsky.social
23.07.2025 11:55 — 👍 4 🔁 2 💬 0 📌 0

Many BPNet models will be made available also correct id is @damlaob.bsky.social
23.07.2025 11:53 — 👍 1 🔁 0 💬 0 📌 0

Next is
@damlaob to talk about putting new Deep learning models of TFs into #Jaspardb #RegSys #ismbeccb2025 @amathelier.bsky.social
23.07.2025 11:50 — 👍 4 🔁 2 💬 1 📌 0

First flash talk session starts with @aryankamal.bsky.social who represents his analysis about understanding transcriptional regulation in blood generation #regsys #ismbeccb2025
23.07.2025 11:44 — 👍 3 🔁 1 💬 0 📌 0

Second proceedings talk by an Panagiotis Alexiou about benchmarking methods for miRNA-target prediction #regsys #ismbeccb2025
23.07.2025 11:23 — 👍 1 🔁 0 💬 0 📌 0

First proceedings talk in #RegSys by Yijie Wang about GRN inference from single cell data
23.07.2025 11:05 — 👍 1 🔁 0 💬 0 📌 0
PhD candidate at Helmholtz Institute for Pharmaceutical Research Saarland
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. https://kiharalab.org/ YouTube: http://alturl.com/gxvah
Fellow of the Kalinina Lab at the Helmholtz Institute for Pharmaceutical Research Saarland (HIPS)
Ph.D. student at the Institute of Computational Genomics @vanvanka123, University Hospital @RWTH Aachen
#timeSeries #epigenomics #probabilisticMachineLearning
Dad, bioinformatician, ex-expat. Currently working on transposon bioinformatics & infectious disease genomics.
Scientist at NHGRI/NIH. Mapping complete genomes, finished a couple, working on more 🐮🐟🐒. Posts are my own opinion.
Associate Professor
DFCI & HMS
Diverse Human References Drive Genomic Discoveries for Everyone
Bioinformatics and Computational Genomics at @HHU.de. Algorithms for pangenomes, structural variation, genome assembly, haplotype phasing, etc.
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
Group Leader - Reader (Assoc. Professor) in AI and Cancer Epigenetics at Blizard Institute, Queen Mary University of London.
#GeneRegulation, #Chromatin, #Epigenetics, #AI, #bioinformatics
https://www.qmul.ac.uk/blizard/all-staff/profiles/radu-zabet.html
Postdoctoral researcher at the Norwegian Centre for Molecular Biosciences and Medicine (NCMBM), University of Oslo
#generegulation #deeplearning
A scientific journal publishing cutting-edge methods, tools, analyses, resources, reviews, news and commentary, supporting life sciences research. Posts by the editors.
Nature Genetics is a monthly journal publishing high impact research in genetics and genomics. Part of @natureportfolio.bsky.social
Repost/like=interesting/relevant, not necessarily endorsement.
A @natureportfolio.nature.com journal on mathematical models and computational methods/tools that help advance science in multiple disciplines. https://www.nature.com/natcomputsci