
🚨New preprint out!
🧬Short reads can now decode centromeres.
🌍We reveal population-scale centromere haplogroups and their links to disease.
biorxiv.org/content/10.1... (1/n)
@adamnovak.graphs.vg
Bioinformatics Programmer at @ucscgenomics.bsky.social working on #pangenomics. http://orcid.org/0000-0001-5828-047X

🚨New preprint out!
🧬Short reads can now decode centromeres.
🌍We reveal population-scale centromere haplogroups and their links to disease.
biorxiv.org/content/10.1... (1/n)
A field in which only the top 5% of participants are able to practice is not a field that can continue to exist.
29.07.2025 17:57 — 👍 4 🔁 0 💬 0 📌 0
A brown circle with a stick behind it and a handle indicating a drum. The drum skin has three stickleback fish with microbes of various shapes and sizes coming from it. The green on the bottom and blue of the water indicates where we think the microbes are coming from
Sharing our lab logo that @ethankocak.com made years ago. The drum is an Inupiaq style drum, part of the storytelling/scicomm we stress in the lab. We study population variation in the stickleback gut microbiome, captured nicely in the design. I get compliments all the time!
27.07.2025 23:13 — 👍 195 🔁 27 💬 6 📌 1
Join HPRC at ASHG 2025 for a workshop on the Human Pangenome: data, tools & workflows.
📅 Oct 14 10AM–12PM | Learn how to access, analyze & work with the pangenome.
Details:
www.ashg.org/product/ashg...
I'm happy to share our latest manuscript "Impediments to countering racist pseudoscience", coauthored with @gillianrbrown1.bsky.social @kztwyman.bsky.social & Marcus Feldman.
25.07.2025 09:17 — 👍 69 🔁 29 💬 1 📌 2
Artistic reconstruction of Late Triassic equatorial Pangaea landscape and paleocommunity preserved in the Pilot Rock White Layer Bonebed of the Chinle Formation in Petrified Forest National Park, AZ, including a pterosaur (Eotephradactylus mcintireae) eating a paleoniscoid-grade actinopterygian fish, stem-turtles, stem-frogs, the skeleton of Revueltosaurus callenderi, and the plant Sanmiguelia lewisii. CREDIT: Brian Engh, LivingRelicProductions.com.
A bonebed in Arizona provides researchers with a glimpse of life in equatorial late Triassic Pangaea. The assemblage includes the earliest documented occurrences of turtles and pterosaurs. In PNAS: www.pnas.org/doi/10.1073/...
25.07.2025 19:53 — 👍 7 🔁 1 💬 0 📌 0
We open our second day at #Hitseq @hitseq.bsky.social with our last key note speaker Tobias Marschall and his insightful talk about the human pangenome and the challenges for structural variation analysis. His work has focused on tackling current limitations such as sample size and remaining gaps
24.07.2025 09:01 — 👍 8 🔁 3 💬 0 📌 0
Two papers in today's issue of @nature.com : 1) we assemble 65 genomes to near completion, including centromeres and the MHC. tinyurl.com/3huhax6w. 2) we sequence 1,019 genomes from the 1kGP with long reads, revealing SVs down to low allele frequencies tinyurl.com/wbx3we9x.
23.07.2025 15:12 — 👍 54 🔁 24 💬 1 📌 2
🚨 We're hiring! NCMBM @ncmbm.bsky.social is looking for 2 new group leaders to join our vibrant research community. Ready to build your own group with strong support and a solid startup package?
Interested and attending #ISMBECCB2025? Let’s connect!
🔗 www.jobbnorge.no/en/available...
Congratulations @benlangmead.bsky.social!
22.07.2025 19:17 — 👍 16 🔁 2 💬 1 📌 0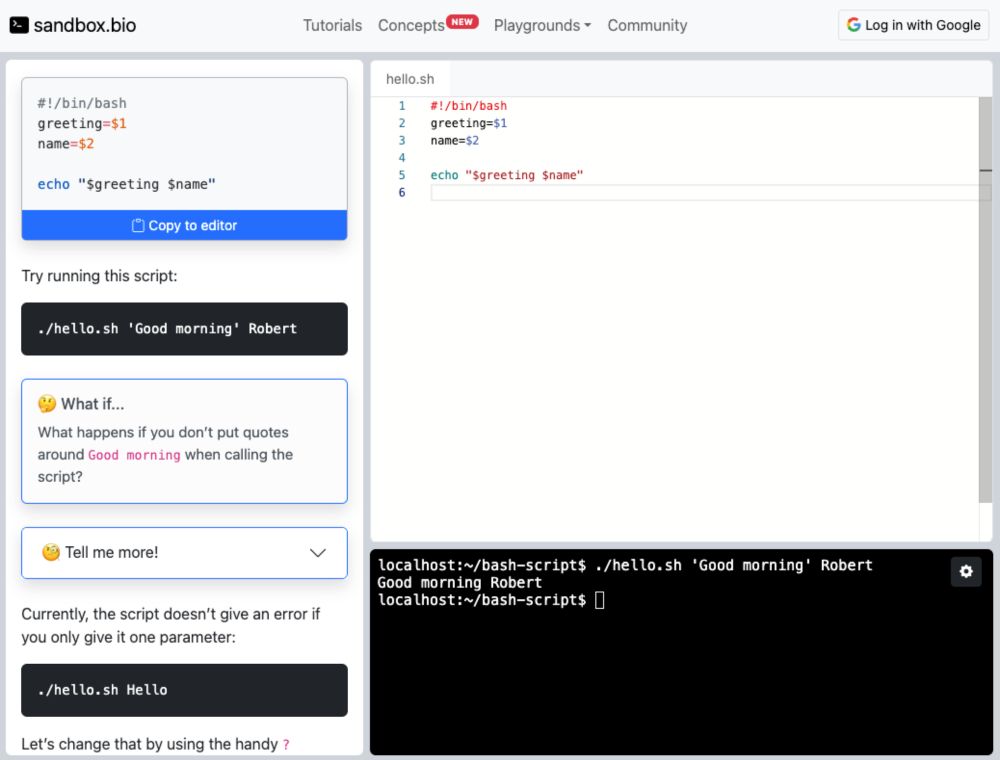
💻 Just released a new tutorial: How to write a Bash script, featuring an IDE and a command-line running directly in your browser. No setup required.
sandbox.bio/tutorials/ba...
📣 JOB ALERT: We are looking for a Senior Software Engineer to join our pathogen genomics team! Be a part of creating and maintaining tools that public health officials are using to track the spread of dangerous pathogens 🦠
Apply here:
careerspub.universityofcalifornia.edu/psc/ucsc/EMP...
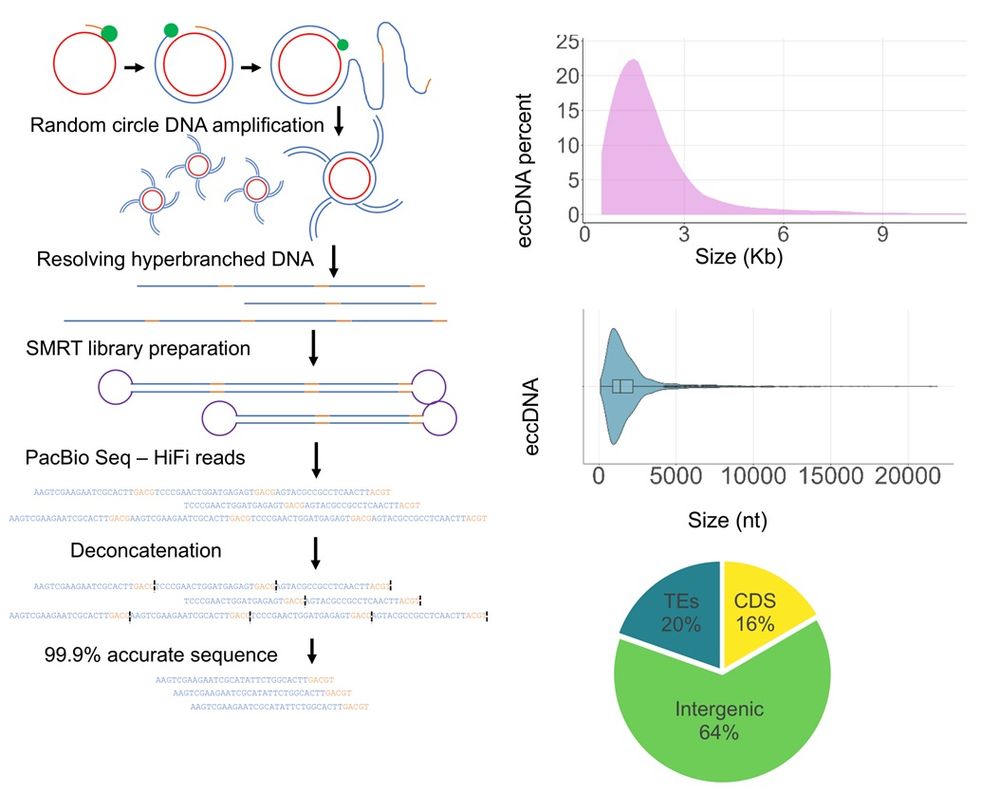
Dynamics and distribution of eccDNA in Arabidopsis. Left: Schematic representation of eccDNA amplification and sequencing via CIDER-Seq pipeline. Top right: EccDNA size distribution in Arabidopsis Col-0 leaf tissues (n = 4,158). The x-axis shows eccDNA size in kb and the y-axis shows the percentage of eccDNAs belonging to each window. Middle right: Size ranges of eccDNAs with marked median and quantiles (n = 4,190). Bottom right: Percentages of eccDNA reads mapping to TAIR CDS, TE, or intergenic databases (values represent the average of three replicates).
Previous studies of extrachromosomal circular DNA #eccDNA in #plants are based on short-read seq. @vanderschuren-lab.bsky.social uses long-read seq to obtain a map of full-length eccDNAs from #Arabidopsis, identifying their genomic origins & epigenetic regulation @plosbiology.org 🧪 plos.io/44Mq3RI
16.07.2025 13:10 — 👍 17 🔁 8 💬 0 📌 0Excited to announce our first interactive article on sandbox.bio, about genomic ranges: sandbox.bio/concepts/gen...
Move & resize the ranges to see how that affects bedtools operations like merge and intersect in real time!
Random paper title generator is here!
{A} {fast,faster,improved} {{algorithm,algorithms} for} {approximate string,pattern} {matching,searching} {using {SIMD,AVX2,AVX512}}

🚨 For everyone who uses All of Us Workbench 🚨
Apparently, if the creator of a workspace no longer has access, *the entire workspace gets deleted*.
Ask me how I know this. 🫠
It doesn't matter who the owners are. Make sure to back up all your work in a workspace with a PI as a creator!
Toil 9.0.0 is out now! You can publish Dockstore workflow metrics back to @dockstore.org out of the box! github.com/DataBiospher...
15.07.2025 18:59 — 👍 1 🔁 1 💬 0 📌 0
My colleague asked me to circulate this job posting for Professor / Associate Professor in Computational Biology / Genomics at the University of Tokyo (P.S. I'm not affiliated):
www.k.u-tokyo.ac.jp/en/informati...

We finished this edition with a delightful keynote by Jean Monlong who guided us through his journey about finding SVs in pangenomes #JOBIM2025 👏🏻
11.07.2025 13:32 — 👍 15 🔁 3 💬 1 📌 1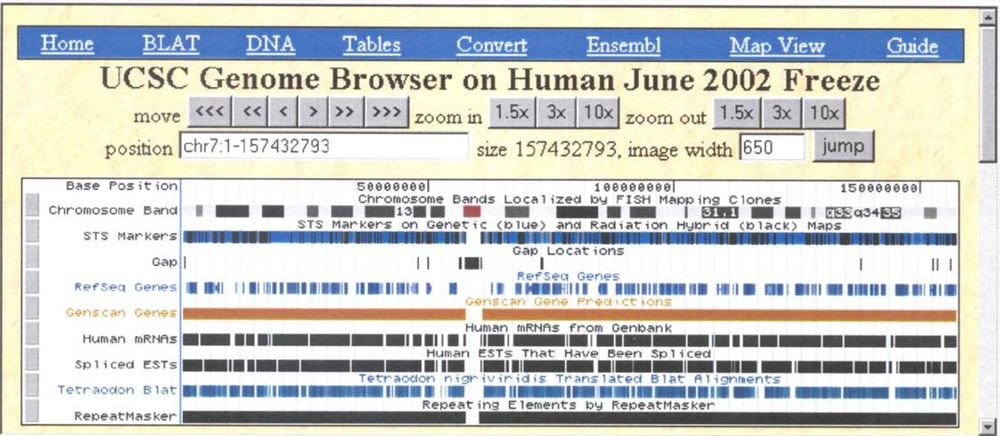
We turn 25 today!
July 7th marks the 25th anniversary of the human genome going online and the start of the UCSC Genome Browser.
Then vs. now, we have 165k monthly visitors, and our codebase is over three million lines of code.
See our news for more: bit.ly/genomeBrowser...


🌍 Hear from Erich Jarvis, chair of the EBP-affiliated Vertebrate Genomes Project at Rockefeller University, as he explains why high-quality reference genomes across the Tree of Life are essential.
👉 bit.ly/401IbWl
@vertebrategenomes.bsky.social @rockefelleruniv.bsky.social @erichjarvis.bsky.social

I'm looking through new archaeal genome assemblies, and some of the species (same genus) show perfect assembly output except in one plasmid - always about 1k different contigs of braided alternate paths but graphs together nonetheless. Is this indicative of some specific biological feature? 🦠🧬💻
03.07.2025 18:44 — 👍 3 🔁 2 💬 1 📌 0
🌱 🧬 New preprint out!
We built and annotated the first reference-free #pangenome of #Arabidopsis thaliana using PGGB (Pan-Genome Graph Builder), spanning 93 assemblies!
Check it out here 👉 www.biorxiv.org/content/10.1...
#Genomics #Bioinformatics

After a quarter of a century, the UCSC Genome Browser remains an essential tool for navigating the genome and understanding its structure, function and clinical impact
https://go.nature.com/40wPxkB

The Secret Rules of the Terminal, by Julia Evans - The cover illustration depicts three people doing arcane terminal magic in a temple with a smoking censer in the background. Each of the three people has curly brown hair and light brown skin. They are all wearing dresses, billowing cloaks, and utility belts with keyboard symbols on them. The one on the left holds a palette of paints and a brush. The one on the right has a staff with a $ symbol on it and a starfish at the top. The one in the centre has a sword and is reading from a book whose cover says “>_” and “./”, which rests on a lectern with a smiling snake wrapped around it.
delighted to announce that my new zine "The Secret Rules of the Terminal" is out today!!
You can get it for $12 USD here: wizardzines.com/zines/terminal
Thanks to new funding from @wellcometrust.bsky.social we will soon be advertising two postdoc positions! Interested? Please get in touch in the first instance and I'll provide more info on the projects. You can also learn more on our lab website www.theradlab.co.uk
#immunosky 🧪
Excited to introduce LiftOn – an open-source tool for accurate, scalable liftover of genome annotations (GFF) across assemblies. 🚀
👉 Code & community: github.com/Kuanhao-Chao...
It’s been incredibly rewarding building this for the genomics community. Can’t wait for your feedback and contributions!
Fantastic work from Anna Kovolenko and team to support the ARCTIC Networks goal to enable portable, low cost, viral sequencing.
The pre-print provides a detailed analysis of alternatives to standard nanopore sequencing library preparation reagents. 👍
🖥️🧬We're thrilled to announce that one of our keynote speakers at #WABI2025 will be the inimitable @benlangmead.bsky.social! wabiconf.github.io/2025/talks/t... Ben's keynote is titled "We are what we index; a primer for the Wheeler Graph era", & it's sure to be a whirlwind tour of full-text indexing!
16.06.2025 12:46 — 👍 20 🔁 5 💬 1 📌 0
Pangenome-aware DeepVariant
doi.org/10.1101/2025...
Comparison of read mapping and variant calling tools for the analysis of plant NGS data
doi.org/10.1101/2020...
#VariantCalling #Pangenomics #Bioinformatics