Rapid epigenomic classification of acute leukemia - Nature Genetics
The authors present a molecular classification of acute leukemia using 5-methylcytosine signatures, together with a neural network-based classifier for clinical use.
Epigenetic diagnosis of acute leukemia within two hours from sample receipt 🚀
Very happy to share the results of a great collaboration between @hovestadt.bsky.social and Griffin Labs, I co-led with @sbenfatto.bsky.social, published today in @natgenet.nature.com
📄 www.nature.com/articles/s41...
🧵1/n
22.09.2025 20:41 — 👍 35 🔁 14 💬 2 📌 3

Introducing ParTIpy, a python package for Pareto Task Inference that scales to large-scale datasets, including single-cell and spatial transcriptomics.
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
15.09.2025 08:39 — 👍 21 🔁 16 💬 1 📌 0

Fluctuating DNA methylation tracks cancer evolution at clinical scale - Nature
Cancer evolutionary dynamics are quantitatively inferred using a method, EVOFLUx, applied to fluctuating DNA methylation.
Studying cancer evolution needs multi-region or single cell seq for phylogenetics, right? Amazingly (I think!) we found single-sample bulk methylation suffices, via analysis of "fluctuating methylation". In @nature.com today led by brilliant @calumgabbutt.bsky.social www.nature.com/articles/s41...
10.09.2025 15:21 — 👍 91 🔁 38 💬 7 📌 2
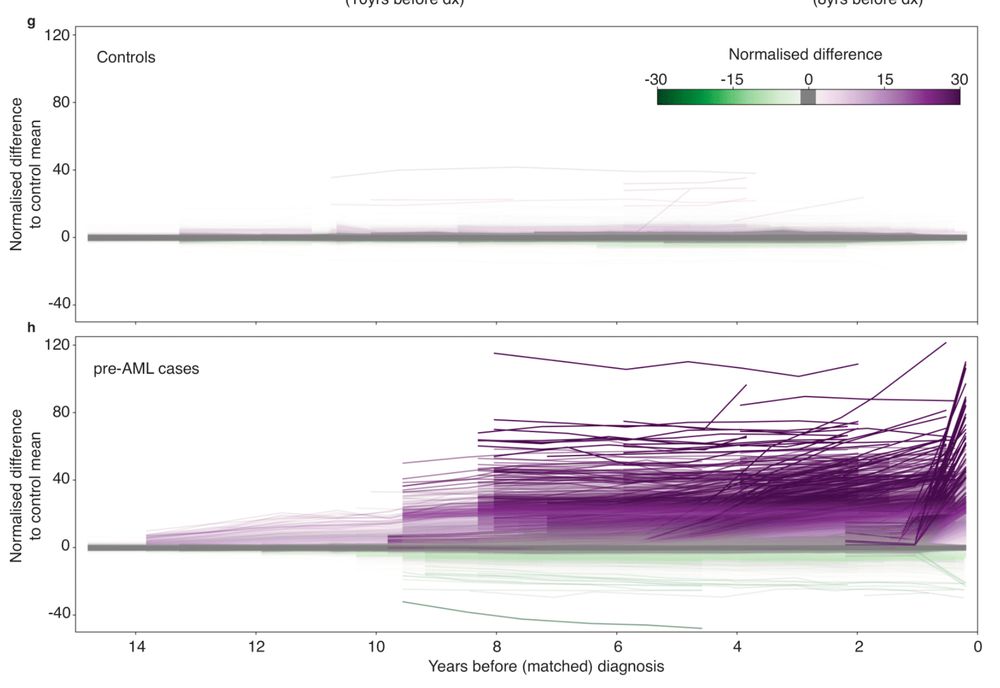
Delighted to share our latest on longitudinal methylation dynamics preceding cancer. Epigenetic signs of AML appear in blood DECADES before Dx.
👉 Early cancer detection
👉 Methylation drivers
👉 Epimutation rates
👉 CpG lineage tracing
www.biorxiv.org/content/10.1...
03.07.2025 20:47 — 👍 22 🔁 13 💬 1 📌 0
Methylation dynamics in the decades preceding acute myeloid leukaemia https://www.biorxiv.org/content/10.1101/2025.06.26.661643v1
28.06.2025 03:35 — 👍 3 🔁 1 💬 1 📌 0
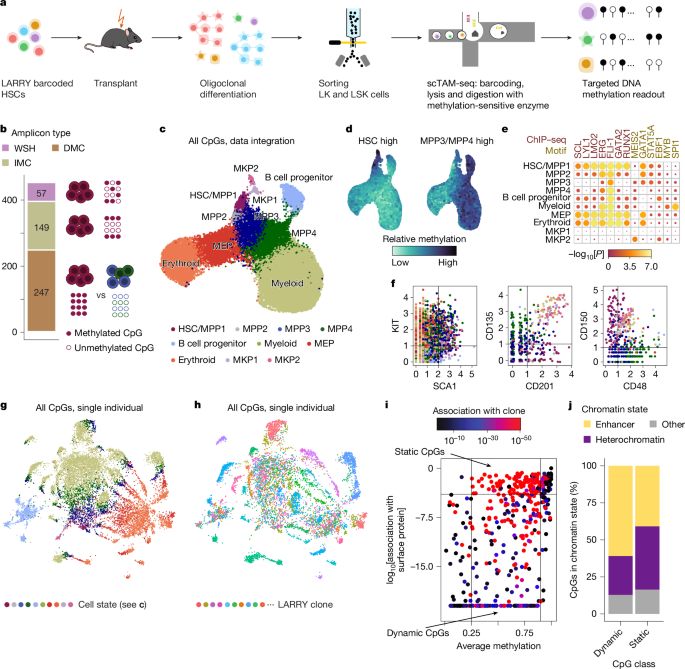
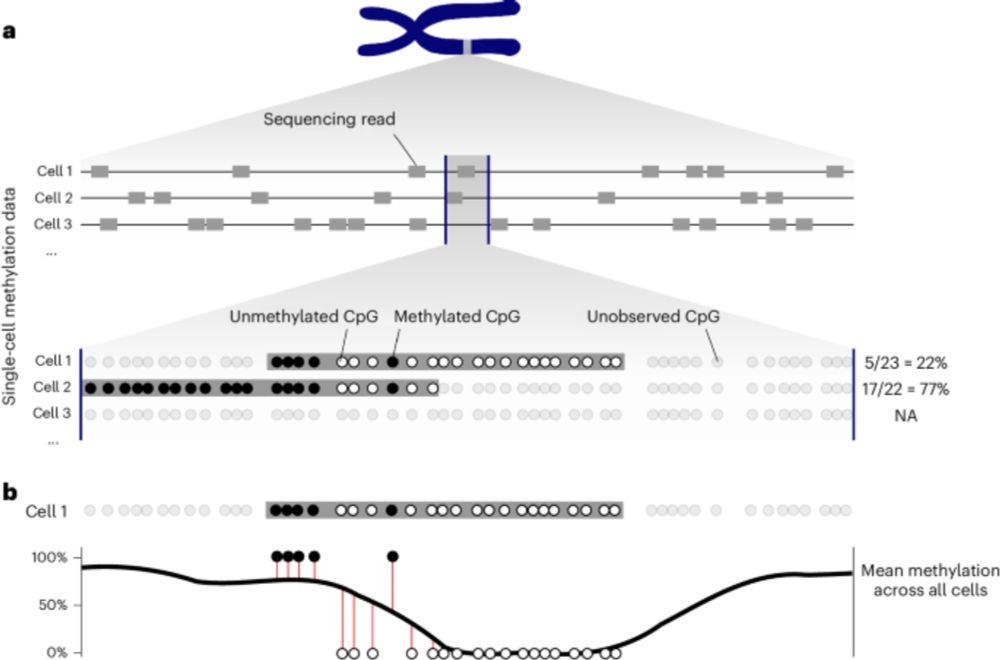
Our work on lineage tracing with somatic epimutations is finally out in @nature.com!
Check out the thread by @larsplus.bsky.social below.
EPI-clone is a revolutionary technique for lineage tracing and it will immediately impact how we trace clones everywhere.
Detailed protocols coming out soon!
21.05.2025 16:06 — 👍 52 🔁 13 💬 5 📌 0
Huge thanks to the rest of the amazing team @scherermich.bsky.social, @indrasingh.bsky.social, Chelsea, and @alejofraticelli.bsky.social
21.05.2025 16:05 — 👍 3 🔁 0 💬 0 📌 0
🚀 Our Nature paper on EPI-Clone is live
I loved analyzing the human data - we discovered that after age 50, blood production shifts to an oligoclonal pattern, and these expansions aren’t driven by CH mutations!
Dive into @larsplus.bsky.social thread for the full story & visuals.
21.05.2025 16:05 — 👍 14 🔁 4 💬 1 📌 0
It is finally out! EPI-Clone is a powerful tool for analyzing clonal dynamics in blood ageing. Thanks to everyone involved, especially @indrasingh.bsky.social, @martinabraun.bsky.social, Chelsea, and @alejofraticelli.bsky.social. Have a look at the thread by @larsplus.bsky.social.
21.05.2025 15:53 — 👍 15 🔁 3 💬 0 📌 0
Bislang gab es keine Methoden, mit denen man sich die klonale Entwicklung von Blutzellen anschauen konnte. Mit EPI-Clone haben wir nun eine solche Methode, die es ermöglicht, klonale Diversität - eine wichtige Eigenschaft eines gesunden Blutsystems - zu untersuchen. doi.org/10.1038/s415...
21.05.2025 15:35 — 👍 5 🔁 1 💬 0 📌 1
🔬‘Barcodes’ written into our DNA reveal how blood ages
Naturally occurring methylation patterns reveal changes in blood which are detectable at age 50 and almost universal by 60
📰 Published in @nature.com by #IRBBarcelona & @crg.eu
Read the news ➡️ bit.ly/4jZHQeB
⬇️ ⬇️
21.05.2025 15:03 — 👍 7 🔁 7 💬 1 📌 0
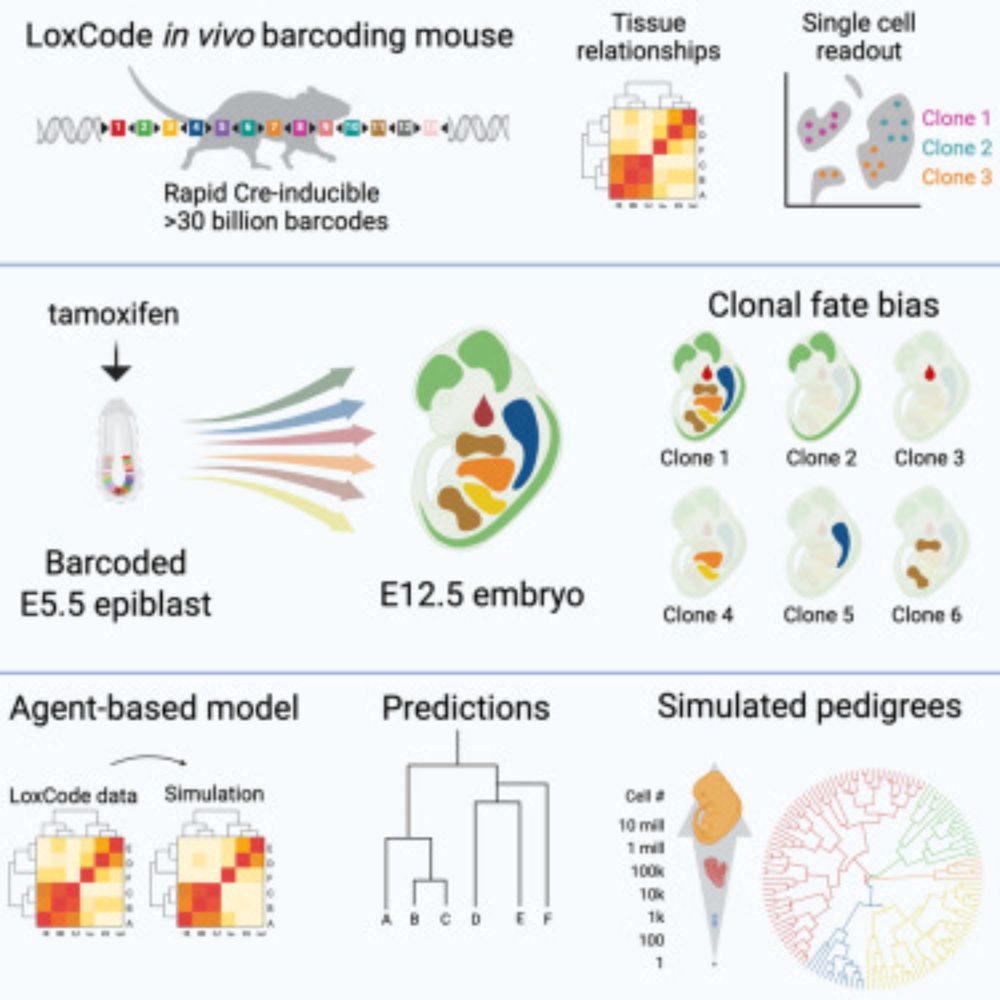
LoxCode in vivo barcoding reveals epiblast clonal fate bias to fetal organs
Much remains to be learned about the clonal fate of mammalian epiblast cells. Here, we develop high-diversity Cre recombinase-driven LoxCode barcoding…
A triumph of perseverance from twitterless Tom Weber, Christine Biben and the team, our in vivo barcoding "LoxCode mouse" used to resolve epiblast fate to fetal organs is finally published in @cellpress.bsky.social and available through @jacksonlab.bsky.social www.sciencedirect.com/science/arti...
16.05.2025 04:32 — 👍 71 🔁 22 💬 8 📌 1

What an inspiring ISCO 2025! Thanks to the organisers & community. Thrilled to have won the 'People’s Choice Best Talk' for my presentation on clonal tracing using epimutations in human hematopoiesis 🌟
Check out our preprint for more. Full paper coming soon! www.biorxiv.org/content/10.1...
14.05.2025 16:45 — 👍 8 🔁 0 💬 0 📌 0
Huge congrats to the whole team for this exciting paper! Can’t wait to see where it takes us!
08.05.2025 17:00 — 👍 8 🔁 0 💬 0 📌 0
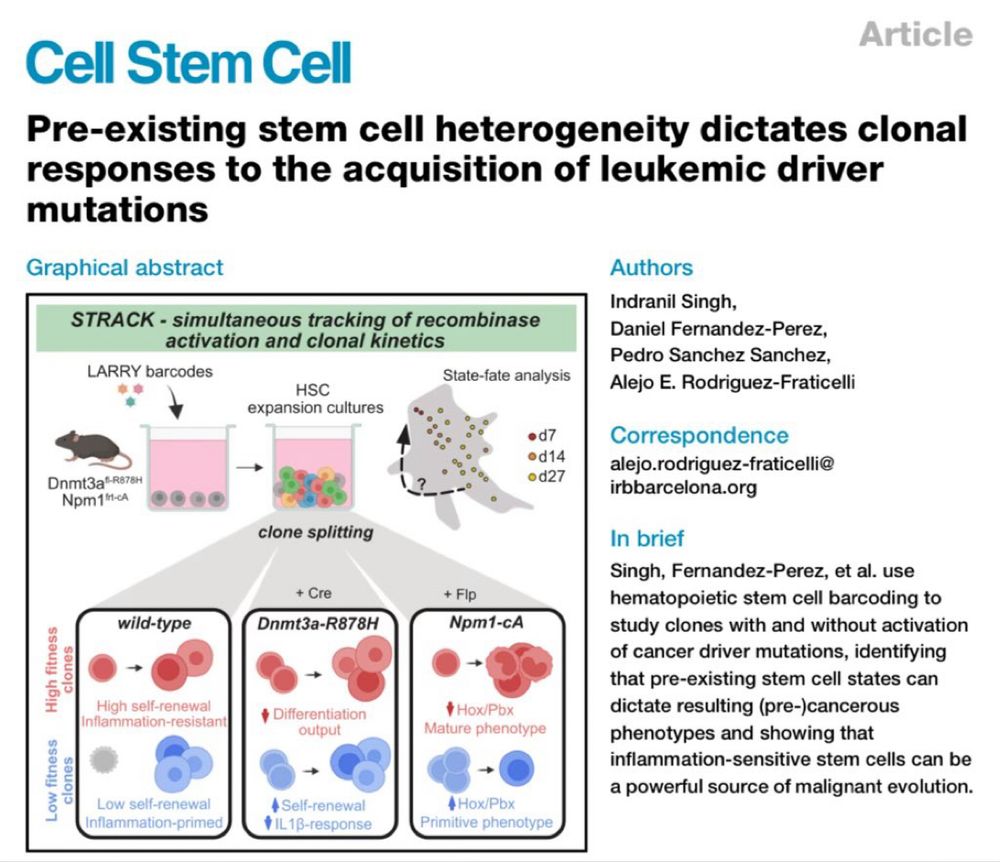
🔥 New Fraticelli lab publication🔥
“Pre-existing stem cell heterogeneity dictates clonal responses to the acquisition of leukemia driver mutations”
Now at Cell Stem Cell, with two new figures, in vivo, and sequential mutagenesis data.
Performed with the support of Cris Cancer and @erc.europa.eu
25.02.2025 16:24 — 👍 109 🔁 26 💬 9 📌 1
Excited to see our EPI-Clone work recognized as a „Method to Watch“ by @naturemethods.bsky.social! Proud to contribute alongside such a fantastic team. Stay tuned for updates!
18.12.2024 16:58 — 👍 6 🔁 0 💬 0 📌 0
GitHub - CompEpigen/epiCHAOS
Contribute to CompEpigen/epiCHAOS development by creating an account on GitHub.
In single-cell epigenomics, comparative analyses have traditionally been performed between cell-type clusters. With epiCHAOS the very talented Katherine Kelly presented the first method to quantify single-cell epigenomic heterogeneity from any type of epigenomic data. doi.org/10.1186/s130... (1/2)
03.12.2024 07:18 — 👍 14 🔁 5 💬 1 📌 0
Thanks for making this!
Would love to be added :)
25.11.2024 09:45 — 👍 0 🔁 0 💬 1 📌 0
Super helpful! Thanks for the great work.
Would love to join
23.11.2024 19:04 — 👍 1 🔁 0 💬 0 📌 0
Thanks for creating this list. Super helpful! Would be great to be added as well.
22.11.2024 08:02 — 👍 1 🔁 0 💬 1 📌 0
Thanks for creating this list, super helpful! 😊
Would be happy to be added as well.
22.11.2024 07:54 — 👍 1 🔁 0 💬 1 📌 0
PhD Student studying human brain tissue using and expanding single-cell genomic methologies. Berlin, Germany
chromatin, transcription, lineage tracing & single cells
News from the Dawson Lab at Peter MacCallum Cancer Centre
Wet and Dry lab scientists interested in circulating tumor DNA and cancer epigenetics
Postdoctoral researcher @EPFL -transcriptional and epigenetic memory- lineage tracing- stem cells- single cells- multiomics
Immunologist and Macrophage plus Barcoding enthusiast 🧬 @UniHeidelberg and @DKFZ
Modelling clonal dynamics in developmental and stem cell biology, @DKFZ, Höfer Group
Just another LLM. Tweets do not necessarily reflect the views of people in my lab or even my own views last week. http://rajlab.seas.upenn.edu https://rajlaboratory.blogspot.com
PI at MSKCC. Synthetic (Developmental) Biology + Molecular Recording + Genomics Tech Dev. Words like physicality of information make my heart go a little faster.
-Scientist interested in Wnt signaling, stem cells and organoids
-Director of pRED/Roche's Institute of Human Biology Basel CH.
-Professor and group leader in Utrecht at the Hubrecht Institute and the Princess Maxima Center
https://www.camlab.ca | ML for cancer genomics + single-cell and spatial computational methods | Toronto Canada 🖥️🧬
Computational biologist interested in cancer evolution, maths modelling and Bayesian stats.
Director, Centre for Evolution and Cancer @cec-icr.bsky.social at @icr.ac.uk
Genomics, epigenetics, evolution, mathematical modelling, data science, bowel cancer
PhD Candidate in Biomedicine @CRG | Lars Velten Lab
PhDing at Barcelona Supercomputing Center | Exploring disease co-occurrences through omics, bioinformatics & HPC — with an eye on AI bias, and occasionally covered in clay.
Incoming group leader @FMI, Basel
Postdoc @MPI-MG, Berlin | PhD @IMBA, Vienna
Investigating cell fate and loss in tissue dynamics
https://www.fmi.ch/research-groups/groupleader.html?group=150
Cell Reprogramming in Hematopoiesis and Immunity, Lund University, Sweden. Tweets by Filipe Pereira (signed FP) and Malavika Nair (for Pereira lab). https://pereiralab.com/
MSCA fellow at @crg.eu w/M. Dias and @jonnyfrazer.bsky.social
> Biological Physics | Proteins | Comp Bio | ML
https://scholar.google.com/citations?user=n55NtEsAAAAJ&hl=en
Engineering cells to understand their decisions
ESPOD Fellow @ebi.embl.org & @sangerinstitute.bsky.social
(Saez-Rodriguez & Parts)
lingering scientist @crick.ac.uk (Briscoe)
Lifelong relationship with microbes, now studying them for a living. Assistant Professor at MedUni Graz. Opinions are my own. He/him 🦠+💻=❤️
More socials at cdiener.com. Lab website at dienerlab.com.
Biochemist 🧪| Empowering Life Scientists and Science Communicators to Achieve Their Career Goals | UCLA PhD + NIH Alum | Pharma R&D @GSK + @AstraZeneca 💊 | #SciComm Director ✍️| Career Coach | Star Trek | Sherlock Holmes | 🎹 | https://larrymillerphd.com