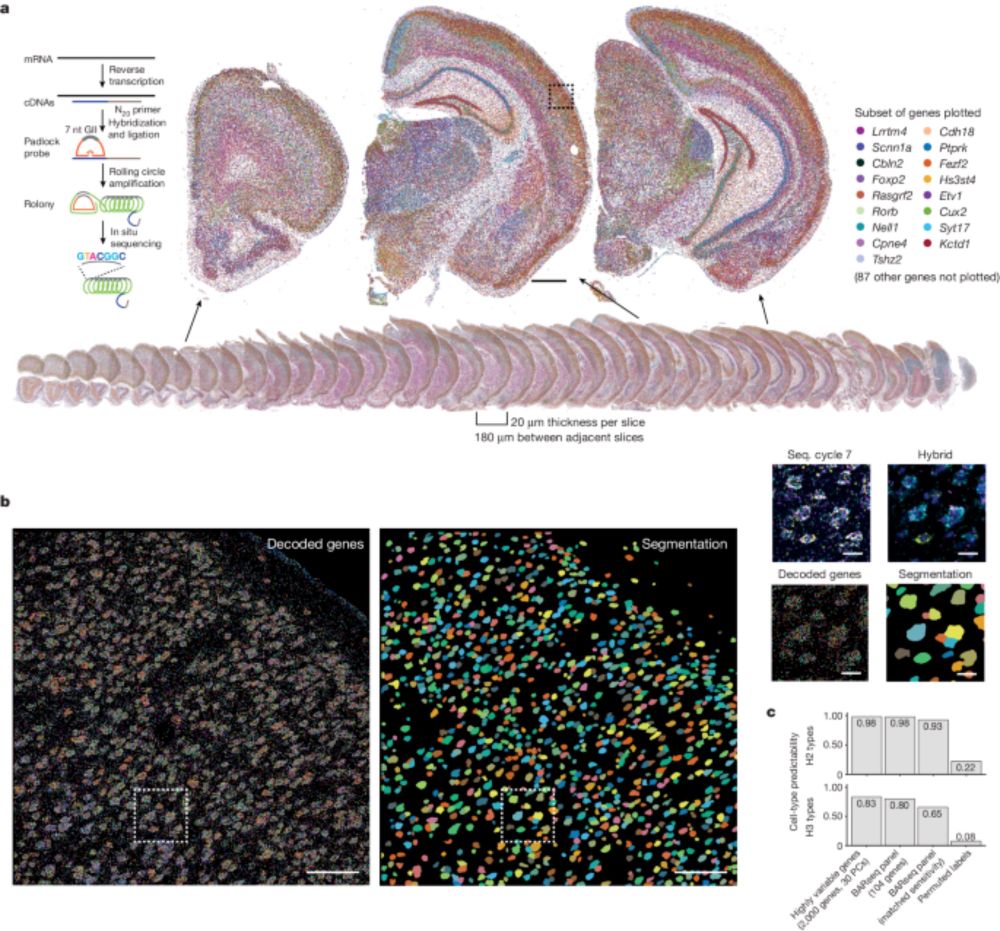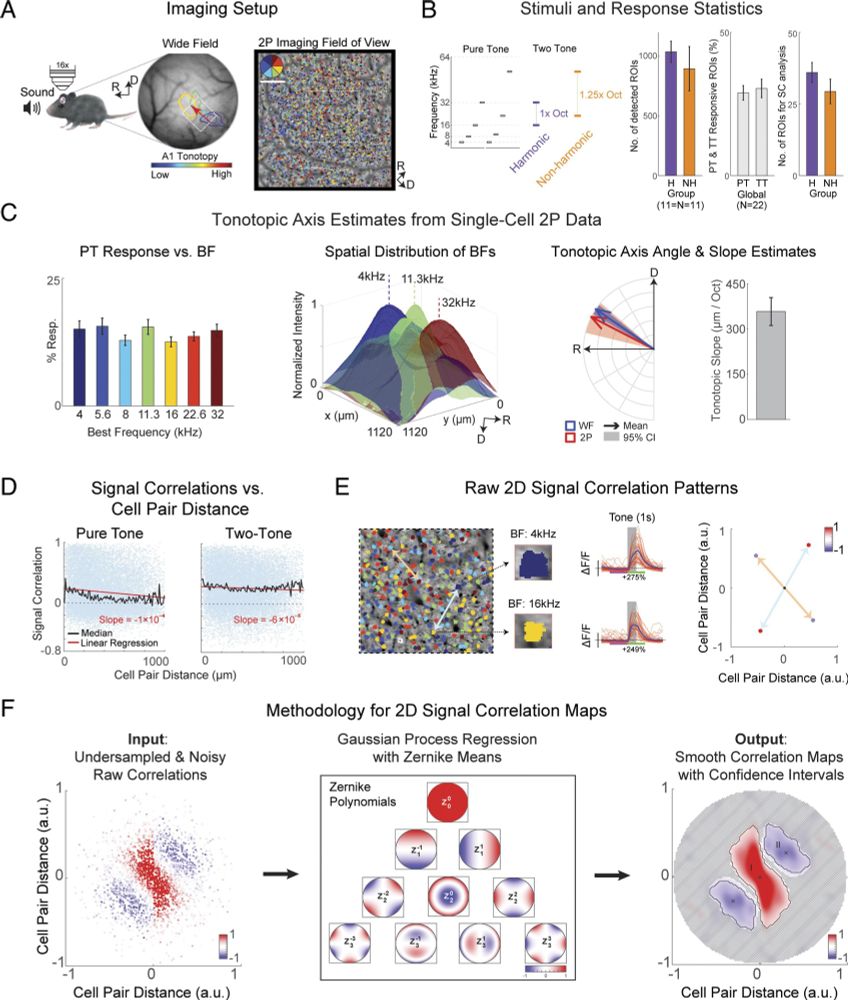
Patchy harmonic functional connectivity of the mouse auditory cortex | PNAS
Analyzing the functional connectivity of the brain is an enormous challenge, as deciphering
functional connectivity requires knowledge of functiona...
New paper out! Peter, Sahar, and Anuthhara show that even though the frequency preference of neighboring neurons in A1 is functionally diverse, the functional connection patterns are functionally precise and harmonically related. Great collaboration with Babadi lab!
www.pnas.org/doi/10.1073/...
11.07.2025 14:25 — 👍 2 🔁 0 💬 0 📌 0
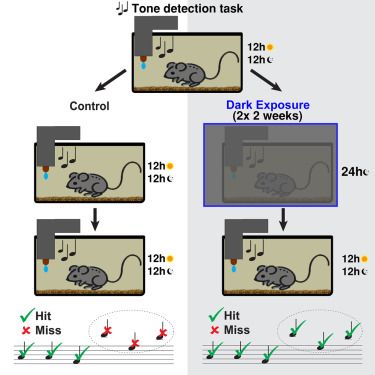
Brief periods of visual deprivation in adults increase performance on auditory tasks
Behavioral neuroscience; Cellular neuroscience; Sensory neuroscience
New paper out: Our prior work had shown that adult visual deprivation can change circuits in the auditory cortex. Now Peter shows that temporary visual deprivation in adults can improve functional hearing and even attenuate age related high-frequency hearing loss: www.cell.com/iscience/ful...
16.11.2024 02:40 — 👍 1 🔁 1 💬 0 📌 0
| bioRxiv
bioRxiv - the preprint server for biology, operated by Cold Spring Harbor Laboratory, a research and educational institution
New preprint that was a long time in the making: HiJee and Travis use in vivo holographic optogenetic stimulation to look at how ensembles in auditory cortex interact:
www.biorxiv.org/content/10.1...
#neuroscience
#auditorycortex
#optogenetics
#Heaing
03.07.2024 01:25 — 👍 4 🔁 1 💬 0 📌 0
Neuroscientist at Stockholm University. 🧠. Our lab studies the neural network basis of innate behaviours, and structure-function relationships in oscillating brain circuits. All views my own. ”Of physiology from top to toe I sing”/W. Whitman.
Professor at Yale School of Medicine; Associate Director of Yale MD/PhD Program; Amateur Perfumer; Retired Attorney; Court-Appointed Special Advocate, CASA-NYC. 🧪🧪🧪 🦅🦅🦅🏈🏈🏈
https://medicine.yale.edu/profile/michael-nitabach/
Glasshalfemptologist.
https://drugmonkey.wordpress.com/
• Neuroscientist • Development • Visual Circuits • Plasticity • BRAIN
Backpacking, fly fishing, quilting.
Skeets are my own. she/her/hers
Med School Professor. Brain Physiology researcher. Biomedical Engineer. Biomedical device work. I stimulate mammalian brains to improve cognition. OMHIWDMB.
We study the functional organization of the auditory system at the University of Illinois at Urbana-Champaign. We are particularly interested in top-down modulation and the impact of aging. Visit at llanolab.com
Development, evolution and diseases of the brain @ Kanazawa, Japan.
金沢で脳の発生・進化・疾患の研究。ラグビーと遺跡めぐり
https://www.facebook.com/hiroshi.kawasaki.372
https://researchmap.jp/hiroshikawasaki?lang=ja
https://orcid.org/0000-0002-2514-1497
The wanderer. The clown. The chairman.
President and CEO, @alleninstitute.org
Opinions are my own.
Professor and Chair of Biomedical Engineering, Boston University. Neuroscientist, Biomedical Engineer, very concerned citizen. Advocate for universal healthcare, housing, pedestrians, cyclists, and transit. he/him
Neuroscientist at CSHL. Interests: neuroAI, molecular connectomics, & cortical circuits. Co-founder of Cosyne and NAISys meetings.
Neuroscientist at University of Bristol. Cortical circuits, PFC, interneurons, dopamine, brain development.
https://www.anastasiades-lab.org
Auditory neuroscientist and fearless leader of the Area 41 Lab @ Pitt. Same handle @vatsun on @neuromatch.social #opentowork.
Neuroscientist. Laboratory of Sensory Perception
Professor and Chairman, Dept. Neurobiology,
University of Maryland School of Medicine
Views are my own
Husband, Father and grandfather, Datahound, Dog lover, Fan of Celtic music, Former NIGMS director, Former EiC of Science magazine, Stand Up for Science advisor, Pittsburgh, PA
NIH Dashboard: https://jeremymberg.github.io/jeremyberg.github.io/index.html
Job: Neurobiology @ USC
Likes: Bikes, crypto, techno
What’s next?
Scottish neuroscientist (Assistant Professor @ University of Pittsburgh). Likes brains, books, maths, and music. Avid endurance runner and cyclist.
rural neuroscientist
caveat emptor: https://bsky.app/profile/dddavi.bsky.social/post/3k5bnogiq5r2j
Theoretical and Computational Neuroscientist at UCSD www.ratrix.org. Neural coding, natural scene statistics, visual behavior, value-based decision-making, statistical & non-statistical inference, history & philosophy of science, research rigor, pedagogy.
UCSD professor of neurobiology and neurosciences
The Union of Concerned Scientists puts rigorous, independent science to work to solve our planet's most pressing problems.
ucs.org