
📢 Please help spread the word: We’re hiring a Head of Student & Postdoc Affairs to coordinate our international PhD program, provide guidance & career counseling, lead training programs, collaborate on EDI initiatives, and manage alumni relations. Apply at www.fmi.ch/education-ca...
04.12.2025 07:23 — 👍 3 🔁 8 💬 0 📌 2
This past month marked the end of an era at the @schierlab.bsky.social @biozentrum.unibas.ch. It was a meaningful, intense, often challenging, but also incredibly rewarding chapter of my scientific journey. Thank you all in the Schier Lab for the unconditional support and the Schierleading!
02.12.2025 13:47 — 👍 29 🔁 5 💬 4 📌 1
Research
IMB Mainz
Lastly, I’m excited to join @imbmainz.bsky.social in 2026 to start my own lab. We'll explore new mechanisms in eukaryotic gene expression, leveraging ‘evolutionary play’ to uncover how regulation, repurposing, and hijacking shape RNA biology. tinyurl.com/y4x29ctt
Thanks for reading! 20/20
19.11.2025 23:21 — 👍 19 🔁 5 💬 0 📌 1
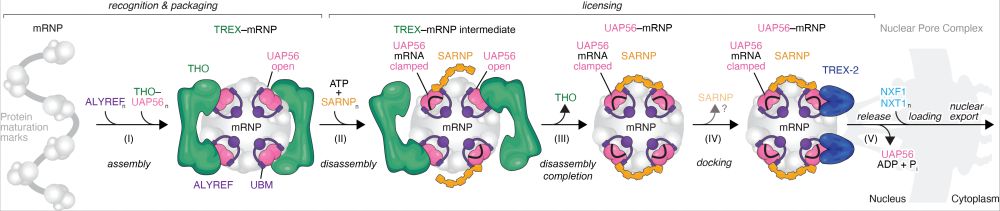
Finally out in @nature.com! We uncovered a mechanistic framework for a general and conserved mRNA nuclear export pathway. www.nature.com/articles/s41.... 1/
19.11.2025 23:21 — 👍 103 🔁 42 💬 3 📌 1
Are you an early-stage graduate student (2nd or 3rd year) or early-stage postdoc based in the US or Canada, working primarily in Drosophila? Would you like to help improve the experience of all trainees working in Drosophila research? If so, read on.
(Please repost to reach a broad audience.)
12.11.2025 04:49 — 👍 74 🔁 150 💬 1 📌 2

New paper alert! Scientists in Clemens Plaschka’s lab at the IMP and @juliusbrennecke.bsky.social's lab at
@imbavienna.bsky.social solved a decade-old puzzle, uncovering how the information molecule mRNA travels from the cell’s nucleus to its periphery. More: bit.ly/4nHcvys
06.11.2025 16:04 — 👍 67 🔁 29 💬 0 📌 1
great to be at the #EMBOMobileGenome meeting in Heidelberg with a wonderful bunch of colleagues, speakers and attendees.
this meeting is a good reflection of the excitement and collegiality in the transposon field.
kindly sponsored by @embo.org
04.11.2025 20:25 — 👍 51 🔁 8 💬 1 📌 1

One clock, two functions: from daily rhythms to development
Scientists at the FMI and the University of California-Santa Cruz have found that similar molecular machineries control daily circadian rhythms and developmental timing. Their work in worms shows that...
Now in @embojournal.org: Researchers in @labgrosshans.bsky.social & UCSC found that similar molecular machineries control daily circadian rhythms & developmental timing—showing that evolution can repurpose core timing systems to coordinate both daily cycles & growth. www.fmi.ch/news-events/...
04.11.2025 08:28 — 👍 11 🔁 6 💬 0 📌 0

Three FMI researchers awarded coveted SNSF Ambizione Grants
Three FMI researchers — Nanci Winke, Arnau Ramos Prats, and Lukas Anneser — have been awarded a prestigious Ambizione Grant from the Swiss National Science Foundation (SNSF). Their work spans from unc...
Three FMI researchers—Nanci Winke, @neuramos.bsky.social & @lukas-anneser.bsky.social—have received @snsf.ch Ambizione Grants! Their projects explore how emotions shape behavior, how the brain responds to anxiety treatments, and how brain states affect learning & memory 🧠 www.fmi.ch/news-events/...
29.10.2025 09:06 — 👍 20 🔁 4 💬 0 📌 0
The Skok Lab is recruiting an experienced computational scientist to lead single-molecule epigenomic and 3D genome analysis. We integrate long-read sequencing with Hi-C/Hi-ChIP, scmulti-omics, and machine-learning to explore chromatin topology, and gene regulation.
23.10.2025 17:10 — 👍 5 🔁 5 💬 0 📌 0

And the next one: dx.doi.org/10.1038/s44318-025-00585-z . Super-fun collaboration with @partchlab.bsky.social & @gotworms.bsky.social to find similarities between developmental and circadian clocks – supported by @fmiscience.bsky.social Facilities and, financially, @snsf.ch and @erc.europa.eu . 1/n
20.10.2025 13:25 — 👍 20 🔁 11 💬 1 📌 1

Mechanisms of Messenger RNA Packaging and Export
The packaging and export of messenger RNA (mRNA) are essential cellular pathways that bridge the nuclear and cytoplasmic phases of eukaryotic gene expression. During their nuclear maturation, mRNAs ar...
what you always wanted to know about the molecular choreography of nuclear mRNA packaging and export.
thought-provoking review by our colleagues (local or in the RNA community).
@plaschkalab.bsky.social
@rupertfaraway.bsky.social
@thezenklusen.bsky.social
www.annualreviews.org/content/jour...
18.10.2025 15:08 — 👍 25 🔁 10 💬 0 📌 0
Congratulations, looking forward to more insights into this really unusual flamenco transcript!
15.10.2025 03:55 — 👍 1 🔁 0 💬 1 📌 0

Paper alert! doi.org/10.1038/s443... - "A scheduler for rhythmic gene expression". We show how 9 txn factors suffice for rhythmic gene expression of thousands of genes with any phase or amplitude in #Celegans larvae (and also look at the tissues where oscillations happen) 1/n
10.10.2025 14:17 — 👍 24 🔁 13 💬 2 📌 1

Just out in @science.org: Together with the lab of @wilhelmpalm.bsky.social, we used sequential in-vitro/in-vivo CRISPR screens to decipher metabolic adaptations in tumors. We find that acidosis is a dominant factor that shapes energy metabolism and stress resilience. www.science.org/doi/10.1126/...
09.10.2025 19:36 — 👍 110 🔁 29 💬 1 📌 3
⚡⚡Excited to announce I'll be starting my lab at the Max Planck Institute of Molecular Genetics (@molgen.mpg.de) in Berlin in December! Leaving sunny California to join a fantastic environment with colleagues who do super cool work.
🔬🦠I'm hiring at all levels! 🔬🦠Check: www.molgen.mpg.de/fueyo-lab
06.10.2025 13:46 — 👍 86 🔁 26 💬 3 📌 2

A human-specific regulatory mechanism revealed in a pre-implantation model
Nature - Genetic manipulation of blastoids reveals the role of recently emerged transposable elements and genes in human development.
Today in @nature.com, we present our work leveraging functional genomics and human blastoids to uncover a human-specific mechanism in preimplantation development driven by the endogenous retrovirus HERVK.
Special thanks to the reviewers whose comments improved our manuscript a lot! rdcu.be/eI3tD
01.10.2025 18:08 — 👍 135 🔁 50 💬 11 📌 5

Lori Passmore
We’re looking for a #postdoc to join Lori Passmore’s group, within a programme aimed at understanding addition of mRNA poly(A) tails
More info: www.nature.com/naturecareer...
Closes 06 OCT
#ScienceJobs #PostdocJobs
18.09.2025 09:36 — 👍 19 🔁 11 💬 0 📌 2

AussiERV 2025
AussiERV: Australia’s first symposium dedicated to endogenous retroviruses. Join us for this one-day virtual event on November 7th!
Do you research endogenous retroviruses (ERVs) or want to learn more about them? 🧬🤔
Consider presenting at or attending AuusiERV - an inaugural online symposium for ERV research in Australia 🇦🇺 (and abroad) 🗺️
Abstracts due 1st October!
events.humanitix.com/aussierv-2025
#ERV #retrovirus
27.09.2025 07:28 — 👍 15 🔁 10 💬 1 📌 0
We’re excited to host the next TriRhena Gene Regulation Club on November 5!
Attendance is free, but registration (by October 20) is mandatory 👇🏽
26.09.2025 08:25 — 👍 6 🔁 4 💬 0 📌 0
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
24.09.2025 21:45 — 👍 114 🔁 52 💬 8 📌 5
My first first-author paper is out!🎉
Here we propose a model where a silencing complex, PIWI*, assembles on target RNAs to recruit effectors and shut down transposon activity.
Huge thanks to the Brennecke and Plaschka labs, especially Julius and Clemens, and all co-authors!
17.09.2025 13:00 — 👍 48 🔁 20 💬 3 📌 1
Laura Lorenzo Orts
IMB Mainz
I am excited to announce that I will be moving to IMB Mainz next year! The Winter call for the IPP PhD program is now open; if you are interested in maternal #mRNA regulation and #translation in early vertebrate development, please apply! Deadline: 16 October.
More info: www.imb.de/students-pos...
15.09.2025 12:13 — 👍 66 🔁 25 💬 3 📌 3
wrong bioRxiv link here, sorry.
the correct link to the collaboration with Torben and Clemens is:
www.biorxiv.org/content/10.1...
17.09.2025 16:55 — 👍 7 🔁 2 💬 0 📌 0

PIWI clade Argonautes are essential for transposon silencing. Without them, animals are sterile due to massive transposon activity.
But how does piRNA-guided target interaction translate into silencing?
PhD student Júlia Portell Montserrat has an intriguing answer
www.cell.com/molecular-ce...
17.09.2025 10:38 — 👍 89 🔁 41 💬 2 📌 4
I am excited to announce that we are looking for a Lab Manager @fmiscience.bsky.social in Basel. If you want to be part of a growing team investigating development and tissue formation, and are enthusiastic about helping set up a new lab, please check out the role:
www.fmi.ch/education-ca...
14.09.2025 11:46 — 👍 28 🔁 27 💬 0 📌 0
scientist | pacifist | transposable elements | evo-devo
Former PhD @trono-lab.bsky.social @ EPFL
Postdoc @ Jachowicz lab @imbavienna.bsky.social
Group Leader - Reader (Assoc. Professor) in AI and Cancer Epigenetics at Blizard Institute, Queen Mary University of London.
#GeneRegulation, #Chromatin, #Epigenetics, #AI, #bioinformatics
https://www.qmul.ac.uk/blizard/all-staff/profiles/radu-zabet.html
Chair, Computational BIology and Medicine Program, Princess Margaret Cancer Centre, University Health Network.
Associate Professor, Medical Biophysics, University of Toronto.
Disclosures: https://github.com/michaelmhoffman/disclosure/
Resident panhandler, http://trichelab.org/
Posts may contain trace quantities of blood🩸, chromatin 🧬, and stats 🧮
CV: https://scholar.google.com/citations?user=AOoIO74AAAAJ
Views expressed are my own (but for the right price they can be yours!)
Molecular evolution, chromatin, archaea and oddball biology. Associate Professor @oxfordbiochemistry.bsky.social Fellow @trinityoxford.bsky.social
Reproductive biologist 🇵🇹 in 🇩🇪
Institute of Reproductive Genetics, CMG, Uni-Münster
PI CRU326 "Male Germ Cells"
DNA methylation, spermatogenesis, ageing, male infertility
Hobby collector, slowest runner in the west
https://linktr.ee/SandraLaurentino
Associate professor at U Manitoba. Research on epigenetics of environmental exposures across the lifespan. Occasional cat photos. Formerly @notthatdrjones
Views my own.
She/Her
Biochemist and structural biologist with a love for chromatin. Group leader at the Hubrecht Institute.
Transposons and epigenetics.
https://brancolaboratory.com/
Scientist and music nerd. All things machine learning and genomics for gene regulation.
Faculty at Max Delbruck Centrum and Humboldt University Berlin
@mdc-berlin.bsky.social
http://www.mdc-berlin.de/ohler
Baritone at www.byrdland.org
CNRS researcher in Marcelo Nollmann's team at Centre de Biologie Structurale (CBS) in Montpellier, France. Microscopist interested in using advanced imaging techniques (Hi-M, FRET, etc) to study 3D genome organization, gene regulation and epigenetics.
Asst Prof McMaster OBGYN #omics | #epigenomics | #bioinformatics | #genomics | #placenta | #preeclampsia | #machinelearning | #cfDNA | #FirstGen She/her 🇨🇦🇦🇺
Come for the science, stay for the corgi pics
Group leader at iGReD Clermont-Ferrand, drosophila & epigenetics, food & mushrooms
https://www.igred.fr/en/team/ero-epigenetic-regulations-ontogenesis/
Postdoctoral scholar in Dr. Ashby Morrison's Lab at Stanford. B.S. at Texas. Cancer Biology PhD at Iowa. Now interested in linking metabolic signaling to chromatin in yeast. Aspirational biochemist. Film photography enthusiast.
Chromatin and cancer research at UNC Chapel Hill. Opinions are my own. Raab-lab.org
Academic. Scientist. Father. Breast Cancer Researcher. Sometimes on TV/Radio.
Blizard Institute, QMUL, London. Working on inter-individual genetic and epigenetic variation of mammalian ribosomal DNA.
Associate Professor | Dad, Husband |Studying the dynamic epigenome with Chromatin Biochemistry, Biophysics, and Single Cell Biology | fission yeast and murine cells | UCSF Microbiology & Immunology | Parnassus
Physician-Scientist | University of Pennsylvania | Pol I, rRNA, Ribosomes, Chromatin, Transcription | Hematopoiesis and Leukemia | 🩸🔬🧪🧬
https://paralkarlab.med.upenn.edu
Exploring RNA & RBPs in cancer: from cell edges to the chromatin. Microscopy lover, proteomics newbie. Likes computer sciences, physics & photography. Also an illustrator and graphic designer for Science and Biology (see: @STEMDORADOscimag)

















