This 🐍 venom made so many cool multi-day live imaging sessions possible 🔬🐟🩸 @chrmosimann.bsky.social @huiskenlab.bsky.social @daetwylerstephan.bsky.social
23.11.2025 21:29 — 👍 10 🔁 2 💬 1 📌 0
Huge thanks to my fellow co–first authors Kevin and Maksim: together we pushed this project forward. 💪 Grateful to Judith and Borhane for support 🙏 and all collaborators @embl.org @biomedizin.unibas.ch @unimainz.bsky.social @tudresden.bsky.social @dkfz.bsky.social @trowbridgelab.bsky.social @ki.se
19.11.2025 20:10 — 👍 0 🔁 0 💬 1 📌 0

🎯 Inflammation and niche remodelling are hallmarks in both CHIP to MDS, underscoring the role of the microenvironment in early myeloid disease. Targeting this stromal-immune remodelling could open new interception strategies.
19.11.2025 20:10 — 👍 0 🔁 0 💬 1 📌 0

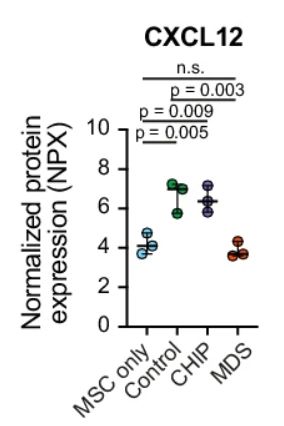
In co-culture experiments 🧫, we showed that healthy & CHIP HSPCs engaged with stromal cells to receive support 🩸🫂(e.g., CXCL12), but MDS HSPCs failed to do so. MDS blasts (!) even triggered an inflammatory program in stromal cells.
19.11.2025 20:10 — 👍 0 🔁 0 💬 1 📌 0

On the immune side 🧪: we found IFN-responsive T cells enriched in MDS that preferentially interact with iMSCs, hinting at a stromal-immune axis of bone marrow niche inflammation. An inflammatory feedback loop?
19.11.2025 20:10 — 👍 0 🔁 0 💬 1 📌 0


Key finding: we identified a novel population of “inflammatory mesenchymal stromal cells” (iMSCs) that appear in asymptomatic and already low-VAF CHIP and expand further in MDS, while healthy CXCL12+ adipogenic stromal cells decline in the bone marrow. 🧬🔬
19.11.2025 20:10 — 👍 0 🔁 0 💬 1 📌 0


We studied human bone marrow samples from 84 donors (age-matched healthy controls, CHIP carriers, and early MDS patients) using single-cell RNA-seq, imaging, flow cytometry, and functional co-culture assays. 🧫
The scale of human stromal niche profiling is what makes this study special! 🦴🩸🧬🔬
19.11.2025 20:10 — 👍 1 🔁 0 💬 1 📌 0
🚨 Excited to share our paper 📢 @natcomms.nature.com
We reveal how inflammation reshapes the human #bonemarrow niche 🦴, home of 🩸 stem cells, in the context of #clonalhematopoiesis and #myelodysplasia
An amazing journey with fantastic colleagues and collaborators, thank you all!
shorturl.at/ad0st
19.11.2025 20:10 — 👍 8 🔁 1 💬 1 📌 1
Huge thanks to my fellow co–first authors Kevin and Maksim: together we pushed this project forward. 💪 Grateful to Judith and Borhane for support 🙏 and all collaborators @embl.org @biomedizin.unibas.ch
@unimainz.bsky.social @tudresden.bsky.social @dkfz.bsky.social @trowbridgelab.bsky.social @ki.se
19.11.2025 20:03 — 👍 1 🔁 0 💬 0 📌 0

🎯 Inflammation and niche remodelling are hallmarks in both CHIP to MDS, underscoring the role of the microenvironment in early myeloid disease. Targeting this stromal-immune remodelling could open new interception strategies.
19.11.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0


In co-culture experiments 🧫, we showed that healthy & CHIP HSPCs engaged with stromal cells to receive support 🩸🫂(e.g., CXCL12), but MDS HSPCs failed to do so. MDS blasts (!) even triggered an inflammatory program in stromal cells.
19.11.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0

On the immune side 🧪: we found IFN-responsive T cells enriched in MDS that preferentially interact with iMSCs, hinting at a stromal-immune axis of bone marrow niche inflammation. An inflammatory feedback loop?
19.11.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0


Key finding: we identified a novel population of “inflammatory mesenchymal stromal cells” (iMSCs) that appear in asymptomatic and already low-VAF CHIP and expand further in MDS, while healthy CXCL12+ adipogenic stromal cells decline in the bone marrow. 🧬🔬
19.11.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0


We studied human bone marrow samples from 84 donors (age-matched healthy controls, CHIP carriers, and early MDS patients) using single-cell RNA-seq, imaging, flow cytometry, and functional co-culture assays. 🧫
The scale of human stromal niche profiling is what makes this study special! 🦴🩸🧬🔬
19.11.2025 20:03 — 👍 0 🔁 0 💬 1 📌 0

Join EMBL as a research group leader in AI in biology. EMBL AI is our new initiative aiming to exploit the full potential of ML/AI-based approaches to advance our understanding of the most complex system in the known universe - life on Earth.
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
(1/3)
09.09.2025 10:06 — 👍 33 🔁 23 💬 1 📌 1

We are all super happy and proud to see our work on the function and evolution of the #cephalic #furrow published in @nature.com. Let me say a few things about the background and history of this work on the #Evolution_of_Morphogenesis (1/12)
04.09.2025 08:21 — 👍 347 🔁 118 💬 16 📌 8

🚨 @dkfz.bsky.social Postdoctoral Fellowships 2025 — Postdoc opening (Mall Lab)
Work at the interface of neurodevelopment, epigenetic repression & cancer neuroscience.
Apply: jobs.dkfz.de/en/jobs/1679...
#Postdoc #Plasticity #Neurodegeneration #CancerNeuroscience
21.08.2025 14:34 — 👍 14 🔁 6 💬 0 📌 1
Looking for a #postdoc opportunity? Funded positions available @cuanschutz.bsky.social to advance our #zebrafish transgenesis tools - and to look into developmental origins of congenital disease!
#devbio
15.08.2025 20:44 — 👍 14 🔁 12 💬 0 📌 0

Excited to share our latest paper @natmethods.nature.com
We present a high-throughput framework to map cellular interactions at ultra-high scale – broadly applicable from whole-organism immune response mapping to personalized therapy response prediction (1/4).
www.nature.com/articles/s41...
07.08.2025 11:24 — 👍 34 🔁 14 💬 3 📌 0

Group Leader - Genome Biology Unit
Are you ready to lead groundbreaking research in Genome Biology? Join us at EMBL! We are seeking a motivated scientist to lead an independent research group addressing exciting and original biological...
To all post-docs: The Genome Biology dept @embl.org
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
30.07.2025 13:41 — 👍 193 🔁 226 💬 0 📌 9
Congratulations Maria with this great story! I'm happy that your visit to our groups at EMBL was so productive and we could support you with the single-cell multiomics experiments. 🧬🧫🧠
31.07.2025 20:49 — 👍 2 🔁 0 💬 0 📌 0
Thrilled to share the story stemming from my PhD!
We used mESC neural differentiation, genome editing and bulk & single-cell approaches to uncover how ZIC2 controls early neural fate by playing a dual and sequential regulatory role as a promiscous pioneer and a selective lineage-specific activator.
24.07.2025 16:27 — 👍 37 🔁 16 💬 7 📌 2
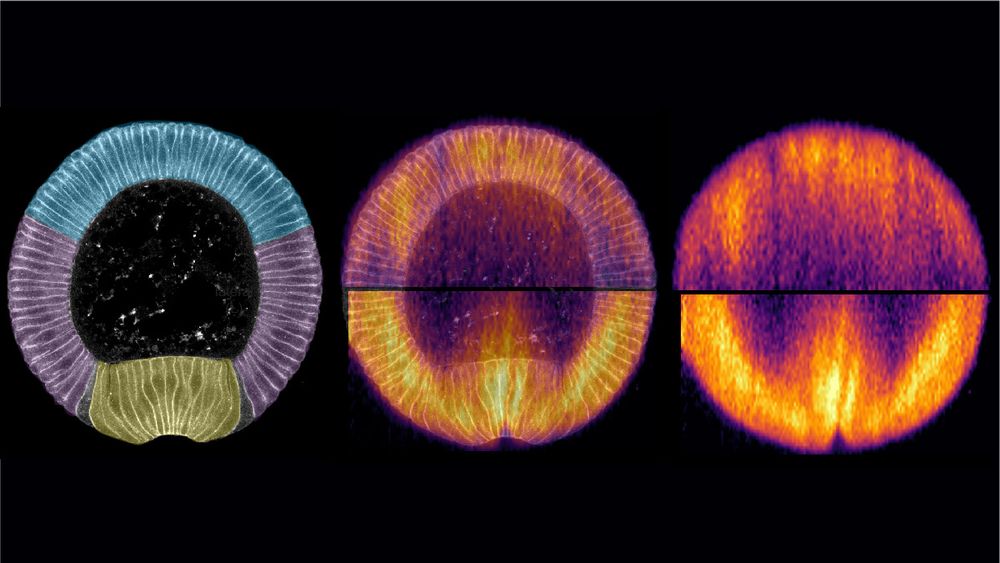
🥁 New article 📢: #Mechanobiology of #development during #Drosophila #gastrulation using #Brillouin microscopy, now in @natcomms.nature.com : rdcu.be/ev6ZX
Collab. w/ @Prevedel_Lab @embl.org , Maria Leptin @marialep.bsky.social, @abhisha-thayambath.bsky.social, Julio Belmonte @ncstate.bsky.social
15.07.2025 14:12 — 👍 52 🔁 20 💬 2 📌 3
Looking for an ultra-high-throughput single-cell RNA+ATAC method? 🎆
Check out our SUM-seq method, now officially out! 🧬
Showcased on macrophages, T cells, and iPSC-derived embryoid bodies; more cell types and tissues to come! 🧫
Reach out for the detailed protocol! 📝 ✨
26.05.2025 10:58 — 👍 4 🔁 0 💬 0 📌 0
Developing integrated technological platforms to study the molecular mechanisms that drive processes in development and disease.
@ Fiolka Lab, UT Southwestern
Developmental biologist interested in neurogenesis, transcription, regeneration, zebrafish, and Middle East geopolitics.
https://bip.ibcs.kit.edu/78.php
https://orcid.org/0000-0003-4411-5646
https://www.germanfishmeeting.org/
Opinions are my own
Assistant Professor at the University of Zurich.
Alumni @schuhlab and @IsabelleVernos.
Cycling biologist.
Lab website: www.reproendo.uzh.ch/en/CavazzaLab.html
Bioinformatician, Department of Biomedicine @ Uni Basel & Swiss Institute of Bioinformatics
Interested in bulk and single-cell omics, data analysis, experimental design, data visualization, R/Bioc, open science, EDI, etc 🧬 🖥️
Herzlich willkommen beim offiziellen Account der #UniMainz!
Web: https://www.uni-mainz.de/
Netiquette: https://presse.uni-mainz.de/social-media/netiquette/
Impressum: https://www.uni-mainz.de/impressum/
Datenschutz: https://www.uni-mainz.de/datenschutz/
🎓 Compte officiel de Sorbonne Université, université de recherche intensive et pluridisciplinaire en lettres, santé, sciences & ingénierie. Suivez notre actualité recherche !
https://www.sorbonne-universite.fr/
Science news from the medical university Karolinska Institutet (KI) in Sweden. Our vision is to advance knowledge about life and strive towards better health for all. https://ki.se/
Postdoc at UCL with James Reading. Previously at EMBL working with Wolfgang Huber. Biostats, R, cancer immunology
Nature Communications is an open access journal publishing high-quality research in all areas of the biological, physical, chemical, clinical, social, and Earth sciences.
www.nature.com/ncomms/
IZKF Research Group Leader at the Institute of Cell Biology (ZMBE) & CeRA, previous Postdoc in Gunawan and Zon Lab - endothelial cells, gonadal function, vascular biology & zebrafish 🐠🔬
Walter Benjamin Fellow at @armilabs.bsky.social Melbourne interested in developmental, regenerative and evolutionary biology. @mpi-bio-fml.bsky.social alumnus. https://marcopodobnik.wordpress.com/
SNSF Postdoc @Stanford Genetics
prev. PhD @EPFL🇨🇭& ESR @ENHPATHY🇪🇺
Comp bio + gene regulation🧬
Stromal Immunology news & views from across the world. Zoe Chua & Anne Fletcher posting/re-posting.
https://keysym.us/KSStromal26
RNAPII &Transcription Regulation -Chromatin Conformation & Nuclear Architecture - ESCs & Neuronal Differentiation - Nuclear Mechanotransduction - Cell Plasticity
UMG, Göttingen, Germany
Chief Science and Strategy Officer, openRxiv. Co-Founder, bioRxiv and medRxiv.
Quantitative stem cell biologist and group leader @TheCrick Institute. Cell decision-making. Cell fate and cell division. @Stanford, @EMBL alumna. ENTP. Mum. Diver.
Leading a research lab with Elena Casacuberta. We aim to unravel how #protists evolved into multicellular animals.
#Multicellularity #Ichthyosporea #holozoans #filastereans #development #diversity #genomes #modelsystems
https://multicellgenome.com
I am a cell and developmental biologist at the Living Systems Institute (LSI) in Exeter. We work on spreading Wnt signals and characterising cytonemes in zebrafish embryos, human neurons, and cancer using high-res imaging and mathematical modelling.
A project of The Center For Scientific Integrity https://centerforscientificintegrity.org/
Daily newsletter http://eepurl.com/bNRlUn
Database http://retractiondatabase.org
Tips team@retractionwatch.com (better than @ replies)
Neuroscience Professor at UKBonn&DZNE Bonn. Bridging the gap between brain and immunity with genomics 🏳️🌈🏴☠️🇨🇳🇹🇷🇫🇷🇳🇪🇩🇪
ORCID 0000-0001-6319-404X


















