YouTube video by Oxford Nanopore Technologies
Exploring Oxford Nanopore sequencing methods for environmental samples
An ONT webinar where I discuss wet and dry lab methods for environmental metagenomics, as well as analysis of 1.5 Tb (!!) of metagenomic sequencing from compost. Featuring the nanoMDBG assembler, strain-dependent AMR profiles, and invertons:
www.youtube.com/watch?v=OZ4P...
17.07.2025 19:24 — 👍 8 🔁 1 💬 0 📌 0
Our guide for best practices in metagenomic assembly and binning of @nanoporetech.com data, highlighting some exciting recently developed tools for high accuracy long reads
21.01.2025 21:26 — 👍 6 🔁 3 💬 1 📌 0
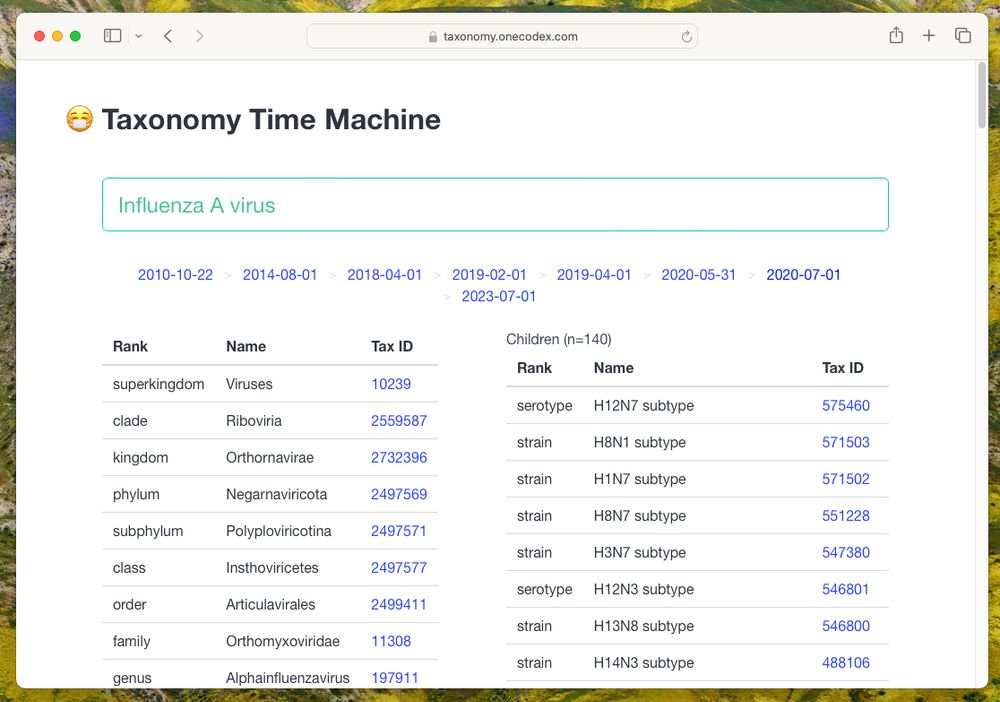
A screenshot of a webpage titled "Taxonomy Time Machine" showing the taxonomic lineage and descendants of the Influenza A Virus
NCBI's Taxonomy changes over time. We built Taxonomy Time Machine to track these changes:
🕰️ app: taxonomy.onecodex.com
📄 pre-print: www.biorxiv.org/content/10.1...
13.12.2024 17:29 — 👍 18 🔁 7 💬 0 📌 1
Long read (DNA, RNA) | Epigenetics (bacteria, human) | Professor of Genomics @IcahnMountSinai | Humid Lab🧫💻🧪 http://fanglab.bio
Working on advancing trust in eDNA at OceanOmics. He/Him.
'It is required that one man should goof gloriously for the people.'
We are a DOE User Facility providing access to integrative genome science capabilities. Located at Berkeley Lab. Work with us: http://jointgeno.me/proposals_B
The Laboratory of Computational Metagenomics led by Nicola Segata
**We are hiring, get in touch if interested!**
http://segatalab.cibio.unitn.it/
Moderator @svetlanaup.bsky.social
PhD candidate//BCMM trainee ambassador
Lynch lab UCSF
studying the human oral microbiome in the context of periodontal disease and health
🪥🦠🫡🏳️🌈
Posts from the NovoaLab members @CRG - Epitranscriptomics And RNA Dynamics Lab. #RNAmodifications #nanopore #ribosome #tRNA #translation #specializedribosomes #inheritance #cancer #machinelearning #discover #imagine #funscience
novoalab.com
Associate Professor, MIT
Still thinking about the 10^9 mutations generated in your microbiome today.
Website: http://lieberman.science
Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org) is an initiative to establish a standardised microbial taxonomy based on genome phylogeny.
Harvard Medical School, Dana Farber Cancer Institute
https://kranzuschlab.med.harvard.edu
Metagenomics, microbes, and molecules.
Advancing how scientists create, use, and reuse microbiome data. Official Bluesky account of the NMDC.
linktr.ee/microbiomedata
Fast, friendly, cutting-edge genome sequencing and bioinformatic analyses for everyone. We help you to sequence smarter, so you can discover more. Live free and seq! 🦠 🧬 🌱🍄🌊
Portsmouth, NH
www.seqcoast.com
Bloomberg Distinguished Professor of BME, CS, and Biostats at Johns Hopkins Univ., tennis player, @StevenSalzberg1 on Twitter, lab: salzberg-lab.org, Substack blog: stevensalzberg.substack.com
Director of bioinformatics at AstraZeneca. subscribe to my youtube channel @chatomics. On my way to helping 1 million people learn bioinformatics. Educator, Biotech, single cell. Also talks about leadership.
tommytang.bio.link
Microbial ecologist: EIC for mSystems Journal, cofounder of http://biomesense.com; President for AMI (http://appliedmicrobiology.org); Father, Scientist, Pilot.
Microbial evangelist and podcaster. Husband, father, moderate. Associate Professor at the University of Puget Sound, facing retirement in June of 2026. Looking for what is next that sparks microbial joy.
Clinic-minded genome + epigenome editor. Professor of Molecular Therapeutics, UC Berkeley. Director for Technology and Translation, Innovative Genomics Institute, ibid.
Beatles fan.
Art work credit: Fyodor Vasiliev "A Meadow in the Rain" (1872).
All views and opinions expressed are my own.
Viral ecology of "brown stuff" 🏞️💩😷. Interested in how viruses impact their hosts in soils, sludge and stool. Particularly fascinated in RNA viruses. Postdoc at UC Davis working on the impacts of wildfire on soil virus communities.

