Client Challenge
Happy to share the published version of this work from my time at Stanford! Here, we leverage mathematical modeling and a large dataset of growth rates, cell sizes, and proteomics to probe how E. coli coregulate their geometry and their macromolecular composition.
doi.org/10.1038/s414...
15.01.2026 03:59 — 👍 5 🔁 0 💬 0 📌 0
It takes two to think - Nature Biotechnology
Nature Biotechnology - It takes two to think
As the scientific pursuit of knowledge faces intimidating challenges, collaboration becomes even more important. I'm a staunch believer in "night-science", which works best when done with others (see this great article from Itai Yanai and Martin Lercher; doi.org/10.1038/s41587-023-02074-2).
25.04.2025 18:10 — 👍 2 🔁 0 💬 0 📌 0
Learning about this problem has been incredibly fun and exciting. I met @akshitg.bsky.social a little over a year ago at APS 2024. Since then, we've worked together on a number of projects. Collaborating with kind and curious people is my favorite part of science and Akshit is a prime example.
25.04.2025 18:10 — 👍 1 🔁 0 💬 1 📌 0
When we combine all known mechanisms, we still can't quantitatively account for the level of strain diversity observed in nature. This suggests a fundamental gap in our ecological understanding that requires new experimental measurements, new theoretical frameworks, and new dialogue between them.
25.04.2025 18:10 — 👍 1 🔁 0 💬 1 📌 0
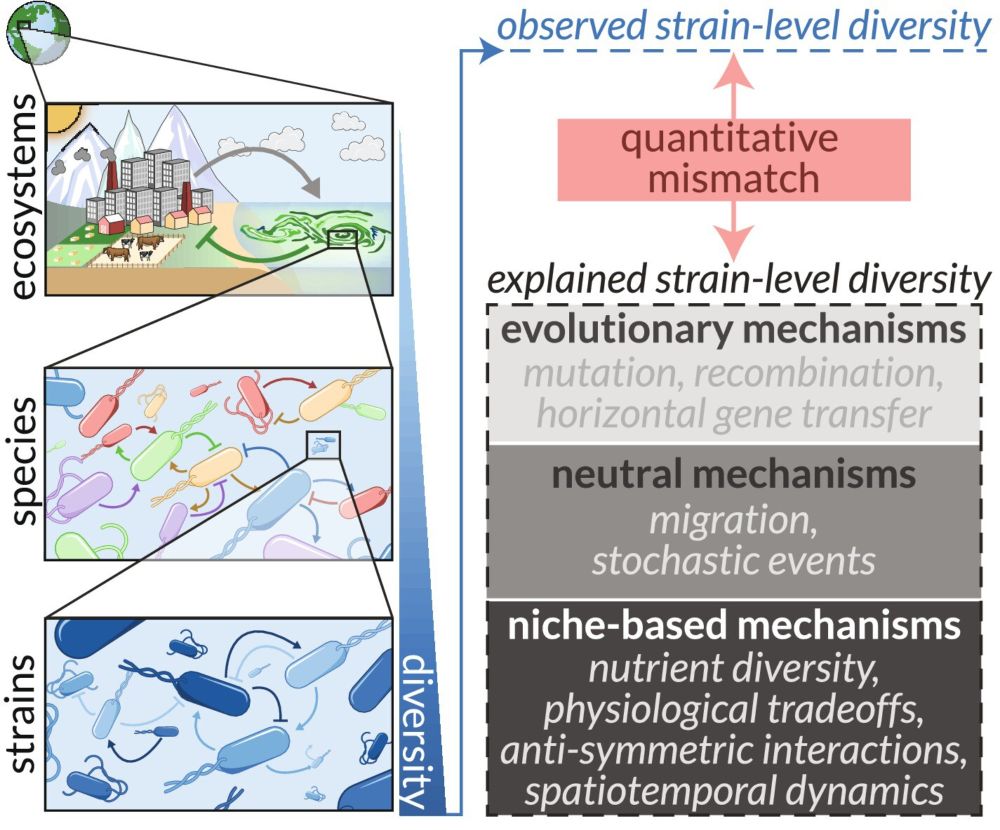
The figure is organized in four sections (A-D), each presenting a different mechanism with its takeaway insight and caveat:
(A) Nutrient diversity: Shows bacteria interacting with different nutrients and metabolites. The takeaway is that strains can develop different nutrient preferences and abilities to consume secreted metabolic byproducts. The caveat notes that strain diversity typically exceeds plausible nutrient diversity, forcing strains to compete within the same niche.
(B) Physiological tradeoffs: Visualizes how different strains allocate their enzyme budget. The takeaway is that tradeoffs in enzyme allocation can yield strains with different metabolic abilities but identical fitness, enabling coexistence. The caveat highlights that any difference in enzyme budgets would alter fitness and disrupt coexistence.
(C) Anti-symmetric interactions: Depicts phage-bacteria interactions. The takeaway is that variation in susceptibility to phage predation can indirectly benefit other community members. The caveat notes that high genetic similarity between strains makes significant differences in susceptibility unlikely.
(D) Spatiotemporal dynamics: Shows bacteria in different spatial compartments. The takeaway is that compartmentalization in space or time can lead to apparent coexistence when averaged across dimensions. The caveat points out that limited space and stochastic temporal dynamics make robust compartmentalization difficult to achieve.
At the top, an overarching question is highlighted: "Niches are not infinitely divisible. How do we quantitatively account for high strain diversity, but limited niche diversity?"
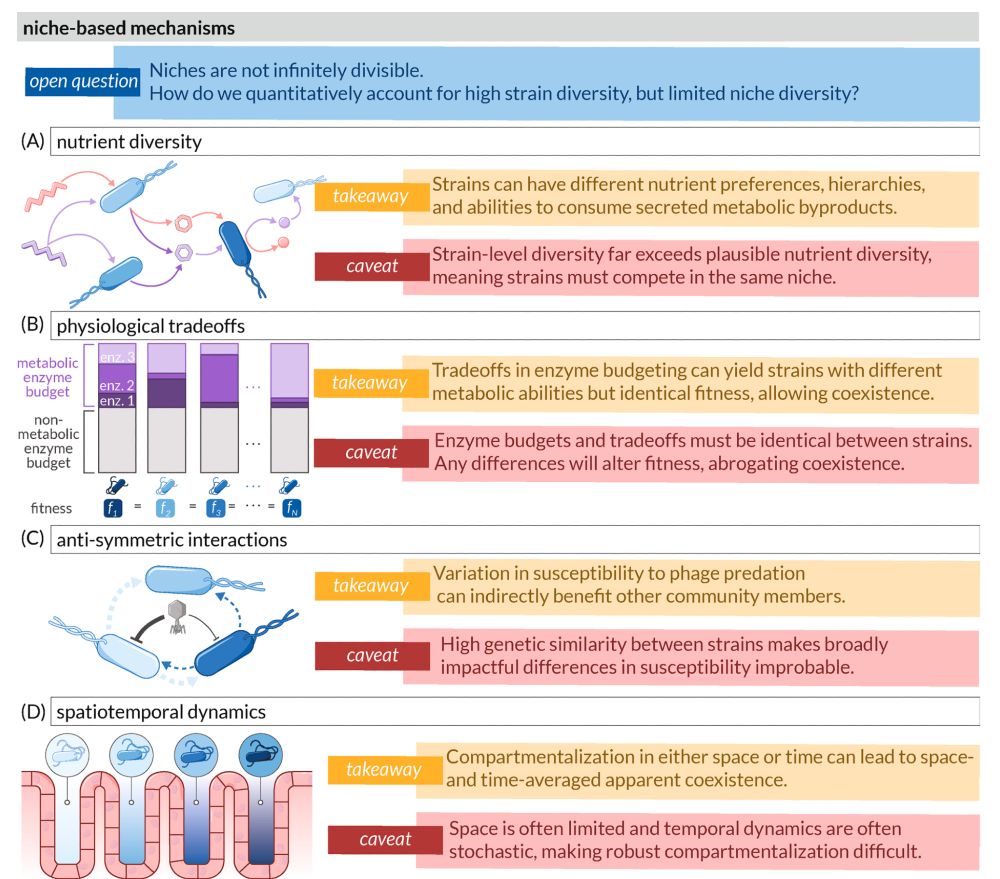
Figure 3 illustrates neutral and evolutionary mechanisms that could contribute to strain-level coexistence in microbial communities.
The figure is organized in two main sections (A-B), each presenting different mechanisms with their takeaway insights and caveats:
(A) Neutral mechanisms: Divided into "migration" (left) showing bacteria moving between spatial compartments, and "stochastic events" (right) depicting regulatory stochasticity, vegetative growth, and spore formation. The takeaway states that dynamics aren't always fitness-dependent as random chance can overwhelm competition. The caveat notes these effects only matter in small population regimes. An overarching question asks at what genetic, spatial, and temporal scales these neutral mechanisms affect coexistence.
(B) Evolutionary mechanisms: Divided into "mutation" (left) showing genetic changes within a lineage, and "horizontal transfer" (right) depicting acquisition of genetic material from other organisms. The takeaway highlights that unlike species, new strains can emerge de novo on ecologically relevant timescales. The caveat emphasizes that strains must originate as single cells and overcome neutral mechanisms to establish. The opening question asks how plastic genomes are over ecological timescales and how widespread horizontal transfer is in nature.
Each mechanism is visually represented with bacterial cells containing simplified genomes, with genetic changes highlighted in red.
We discuss several potential mechanisms that maintain this diversity and highlight their limitations: (1) niche-based (nutrient specialization, physiological tradeoffs, phage interactions, spatiotemporal dynamics), (2) neutral (migration, stochasticity), and (3) evolutionary (mutation, HGT).
25.04.2025 18:10 — 👍 1 🔁 0 💬 1 📌 0
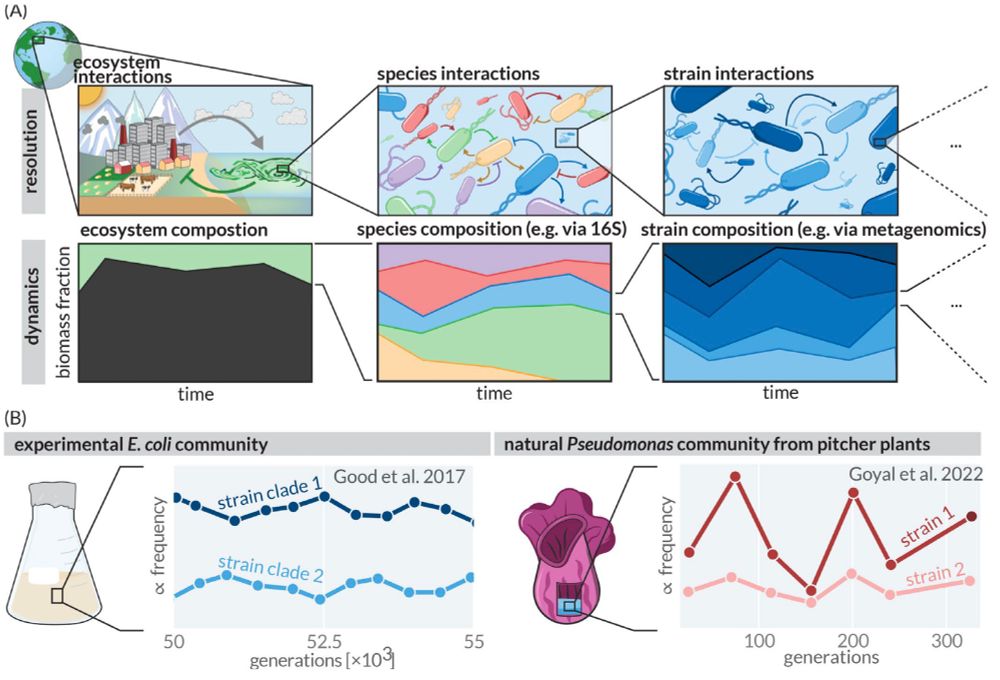
Figure 1 illustrates the concept of coexistence across taxonomic scales.
Panel A shows a hierarchical view of ecological interactions and community dynamics at three levels of resolution. The top row depicts schematic interactions (shown with arrows) at ecosystem, species, and strain levels, zooming in progressively from left to right. The bottom row shows corresponding temporal dynamics of biomass composition at each level, with colored areas representing relative abundance over time.
Panel B provides two specific examples of strain coexistence: Left shows an experimental E. coli community (from Good et al. 2017) with two strain clades persisting over 50,000-55,000 generations in a laboratory flask. Right shows a natural Pseudomonas community from pitcher plants (from Goyal et al. 2022) with two strains coexisting over 300 generations with fluctuating but persistent relative frequencies.
The figure demonstrates how diversity and coexistence patterns exist at multiple biological scales, with particular focus on strain-level diversity that represents the "paradox of the sub-plankton" discussed in the paper.
We call this the "Paradox of the Sub-plankton" - how do organisms with >99.9% genetic identity avoid competitive exclusion and maintain diversity?
25.04.2025 18:10 — 👍 0 🔁 0 💬 1 📌 0
The microbial 'Paradox of the Plankton' asks why many species can coexist on limited nutrients. While we have some understanding of how this work at the species level, another puzzle exists at a finer scale: within species, nearly genetically identical 'strains' show distinct ecological behaviors.
25.04.2025 18:10 — 👍 0 🔁 0 💬 1 📌 0
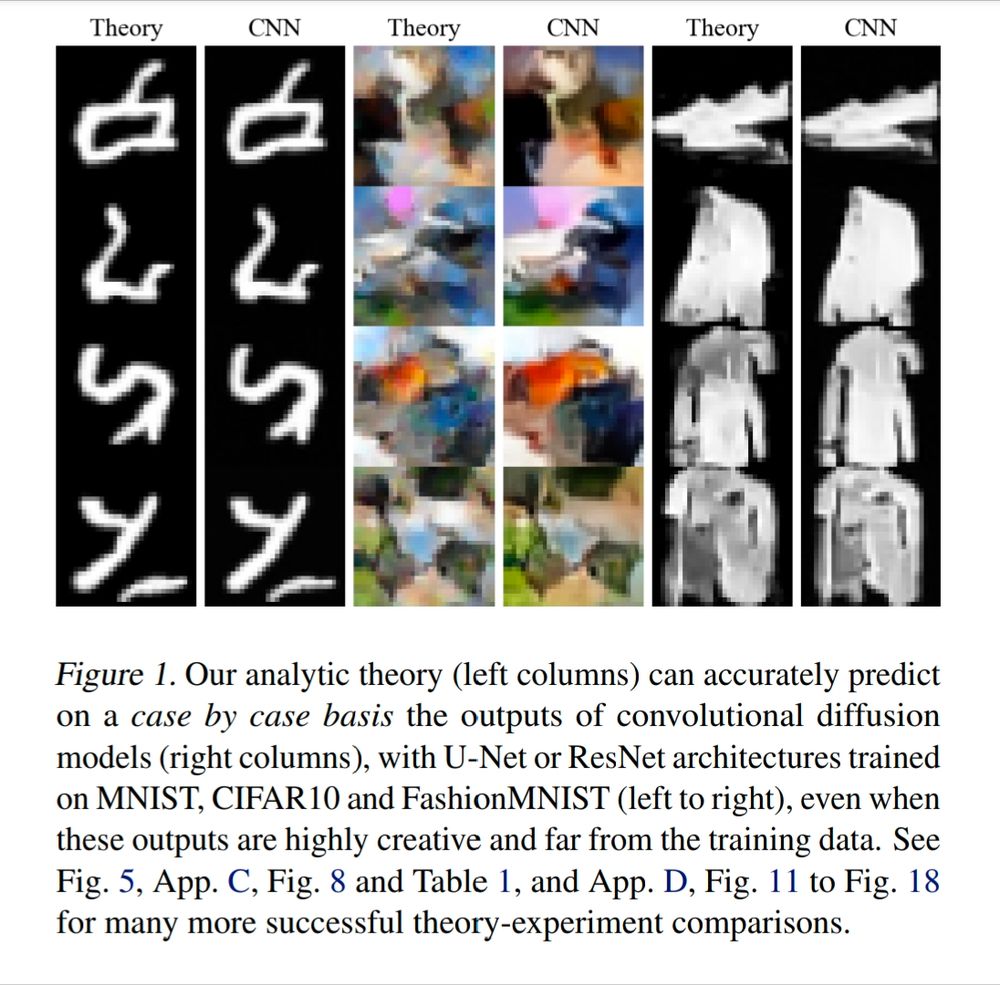
Excited to finally share this work w/ @suryaganguli.bsky.social Tl;dr: we find the first closed-form analytical theory that replicates the outputs of the very simplest diffusion models, with median pixel wise r^2 values of 90%+. arxiv.org/abs/2412.20292
31.12.2024 16:00 — 👍 106 🔁 20 💬 3 📌 8
Generate random draws from a Cauchy just to remind yourself that you almost never actually want a Cauchy anywhere in a probabilistic model.
13.11.2024 14:25 — 👍 19 🔁 4 💬 2 📌 0
SKLAB Home
I am cautiously optimistic that my lab will have an open postdoc position in 2024. If you are potentially interested, please reach out. Or if you know somebody who might be interested, please connect us. My email is on our lab website sklab.science
17.10.2023 01:12 — 👍 10 🔁 9 💬 0 📌 2
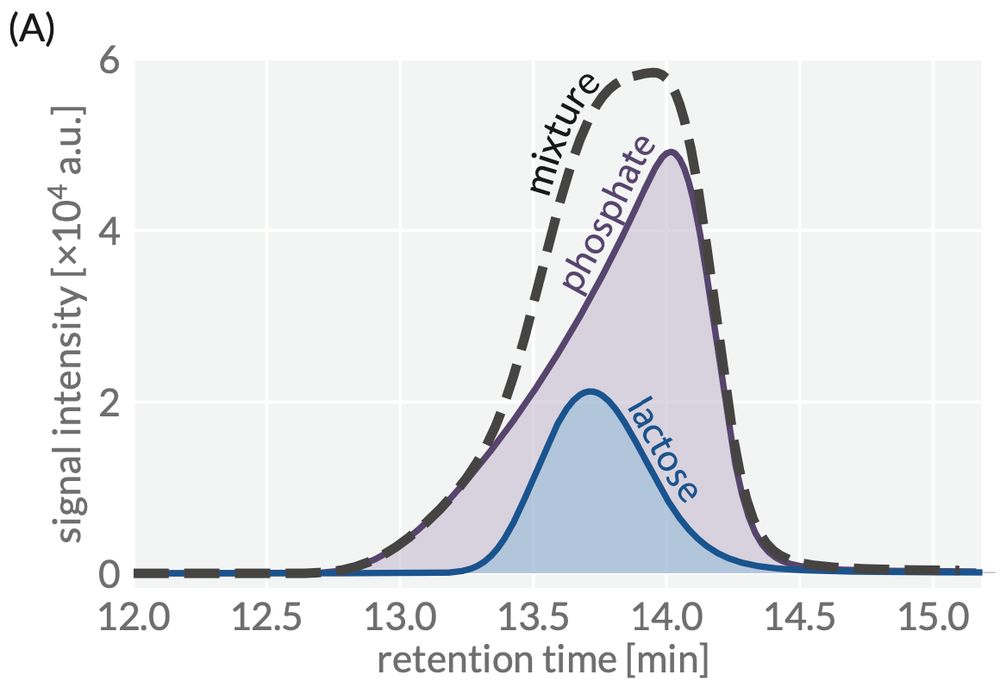
Unlike extant chromatographic processing software, hplc-py allows the user to constrain specific parameters of the fit distribution. This allows one to deconvolve completely overlapping signals like the following:
10.10.2023 16:51 — 👍 0 🔁 0 💬 1 📌 0
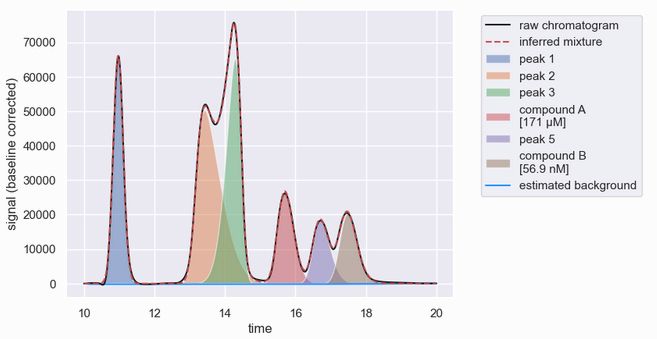
This programmatic interface i) automatically detects peaks in a chromatogram, ii) fits a plausible mixture model to reconstruct the observed signal, and iii) computes and returns properties of each analyte. With hplc-py, you can go from raw data to this in ≤10 lines of Python.
10.10.2023 16:50 — 👍 2 🔁 0 💬 1 📌 0
Over the past two years, I've used HPLC to measure carbon turnover in microbes. The most painful part was quantifying the resulting chromatograms. Today, I (with Jonas Cremer) released hplc-py: A python tool that makes quantification easy. cremerlab.github.io/hplc-py
10.10.2023 16:49 — 👍 11 🔁 3 💬 1 📌 0
If you’re interested in comprehensive guidance for improving your communication skills I cannot recommend the book “Trees, Maps, and Theorems” enough, www.principiae.be/X0100.php. 2/5
30.08.2023 18:20 — 👍 5 🔁 3 💬 1 📌 0
Weather and climate scientist focused on extreme events like floods, droughts, & wildfires on a warming planet. www.weatherwest.com
Reporter covering the Trump White House & 2026 midterms. Emergency backup hockey writer. FR/EN 🇫🇷
jakelahut.com
(Signal: Leak2Lahut.26)
Microbiology editor at a multidisciplinary open-access journal.
Born & raised in Asturias, Spain.
In the UK since 2008.
Dad ×3, husband ×1.
Posts in English and Spanish.
LinkedIn: http://bit.ly/CesarSanchez
The views expressed are my own.
Pues eso.
Choose a better internet where privacy is the default. Start protecting your personal information online with Proton Mail, Proton VPN, Proton Drive, Proton Pass, Proton Wallet, and Lumo.
i write the data-driven politics newsletter Strength In Numbers: gelliottmorris.com/subscribe
wrote a book by the same name wwnorton.com/books/Strength-in-Numbers
polling averages at @fiftyplusone.news
formerly @ 538 & The Economist. email, don't DM, me
Mapping how chemistry perturbs biology to build a virtual model of the human cell.
www.tahoebio.ai
Reporter lady @ms.now
Signal username: brandyzadrozny.53
Email if you must, from a non-work account: brandy.zadrozny@ms.now
Biologist interested in how embryonic development evolves #EvoDevo • Assistant Professor at UFMG • Website: https://brunovellutini.com • Pronouns: he/him
Assistant Professor of Physics at University of Florida. Interested in the evolution and spatial dynamics of microbial communities.
Computer scientist on a life-science mission. Working on pushing bioimage analysis, computer vision, AI/ML methods for the life sciences. Love running and pottery.
Combining frontier AI & frontier biology to help scientists cure or prevent disease
journalist / founder @ thehandbasket.co
email: marisa@thehandbasket.co
signal: https://signal.me/#eu/VssgH88q6WQu7MtH5wF-08JdgWh4iAPWD13eXiOcXQNGdZUXijJBZInD-UtLJKFG
venmo: venmo.com/u/Marisa-Kabas
ko-fi: https://ko-fi.com/marisakabas
Assistant Professor of Physics & Astronomy @ USC
Active Matter + Physical Bioenergetics
https://www.peterfosterlab.com
I'm a theoretical physicist at Durham University
Theoretical Physics Ph.D., Computer Science Teacher. Author of “Intelligenza Artificiale” Zanichelli 2022.
I draw using mathematics and Python, no generative AI.
https://profconradi.com
https://x.com/S_Conradi
At KITP on the UC Santa Barbara campus, researchers in theoretical physics and allied fields collaborate on questions at the leading edges of science.
www.kitp.ucsb.edu
Associate Professor 🧑🔬🧑🏫 at McGill University 🇨🇦 CRC in Spatial organization of living systems 💧🦠🪱🔬
https://weberlab.ca/
Assistant professor in Cell and Developmental Biology at UCSD
@ucsandiego.bsky.social. Loves biophysics, cell mechanics and motility, parasite biology.
Professor, Molecular & Cellular Biology, Harvard University
We are the Cluster of Excellence Physics of Life (PoL) at Technische Universität Dresden @tudresden.bsky.social @dfg.de
Our goal is to unravel the principles underlying the organization of living matter.
https://physics-of-life.tu-dresden.de