
Learn more, use our models, or work directly with us!
Blog: www.profluent.bio/showcase/pro...
Github: github.com/Profluent-AI...
Platform access: docs.google.com/forms/d/1Pdk...
@jeffruffolo.bsky.social
Protein Design / ML @ Profluent Bio | Molecular Biophysics PhD @ Johns Hopkins

Learn more, use our models, or work directly with us!
Blog: www.profluent.bio/showcase/pro...
Github: github.com/Profluent-AI...
Platform access: docs.google.com/forms/d/1Pdk...
We’re incredibly optimistic about the opportunities to solve important, hard problems in protein design by scaling up our models and data. We’ve already ~10x our data scale since training ProGen3, so this really is just the beginning.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0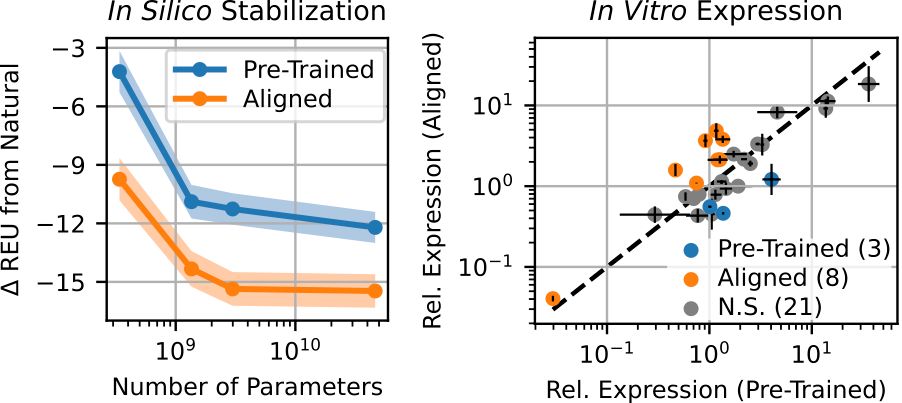
Not only do we see compelling benchmark performance, but also that these aligned capabilities extend to generative settings, which is what really matters for design. Meaning, with just a bit of data we can steer the models to generate the high-fitness sequences we want.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0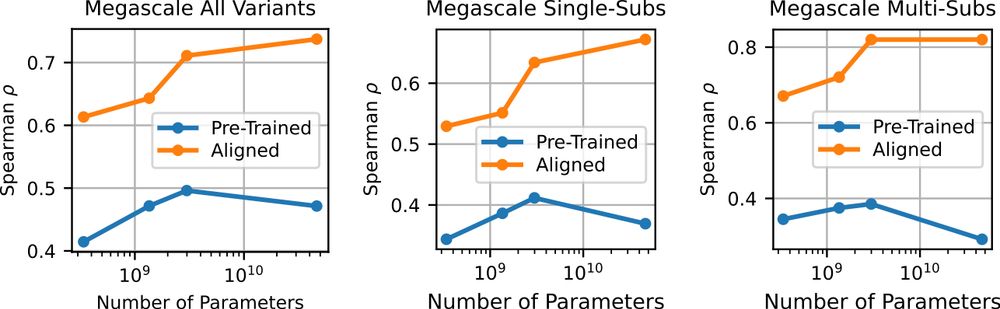
Coming back to fitness prediction, we wanted to see if this greater understanding of protein sequence space translated to stronger predictive power. We turned to alignment, where we use a bit of experimental data to tilt the model towards properties we care about, like stability.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0We think this is the beginning of a new, more meaningful way of understanding what it means to scale protein language models, going beyond ranking of mutations or predicting structural contacts. This will be incredibly useful in shaping how we apply models like ProGen3.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0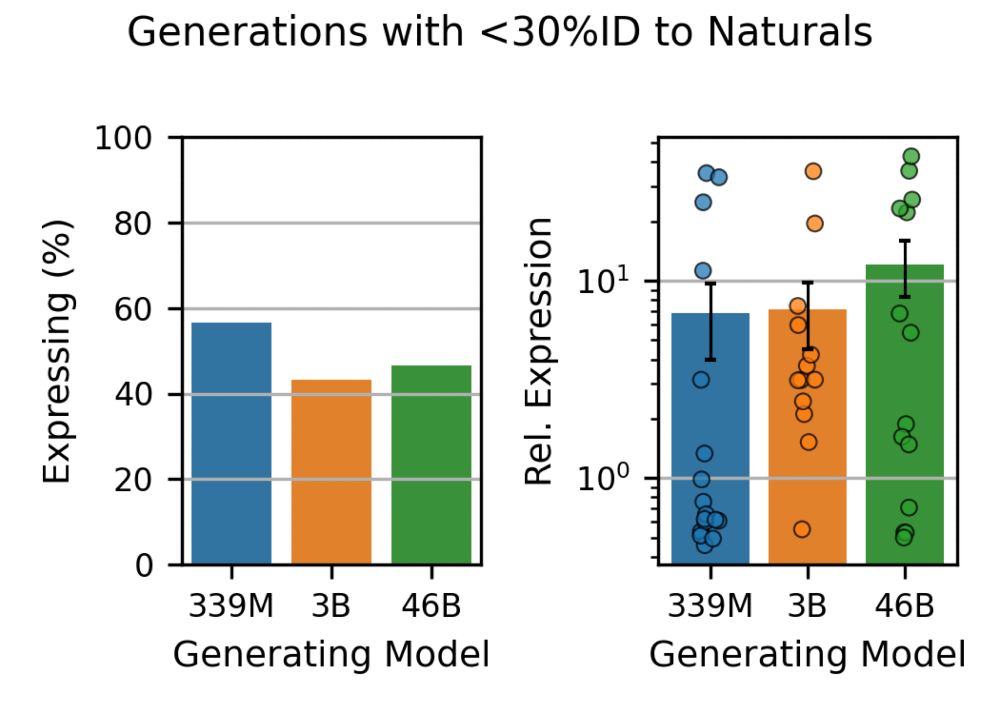
This extends even to proteins that had low (or no) homology to anything in the models’ training data, where we still see comparable rates of protein expression, including for proteins with very low AlphaFold2 pLDDT.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0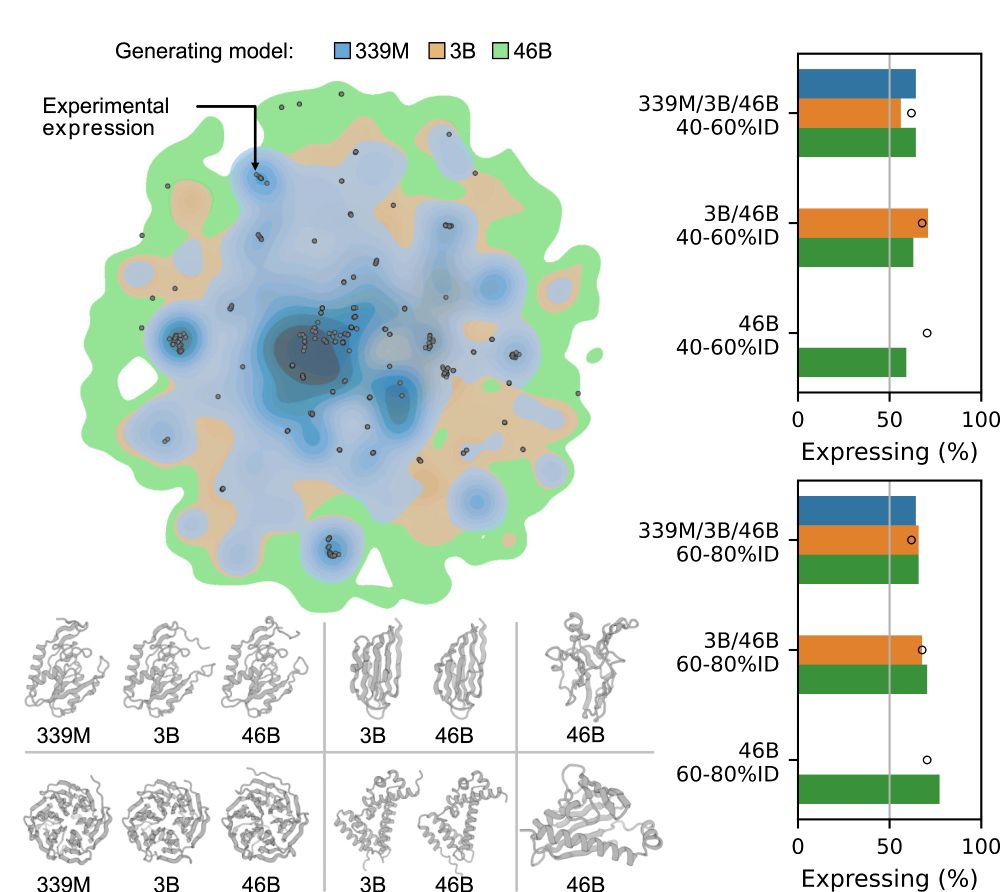
To put this to the test, we experimentally tested the viability (expression) of hundreds of proteins in the lab, and found that this added diversity is real. Generated proteins are as viable as natural proteins, and larger models can come up with more and more of them.
17.04.2025 20:28 — 👍 1 🔁 0 💬 1 📌 0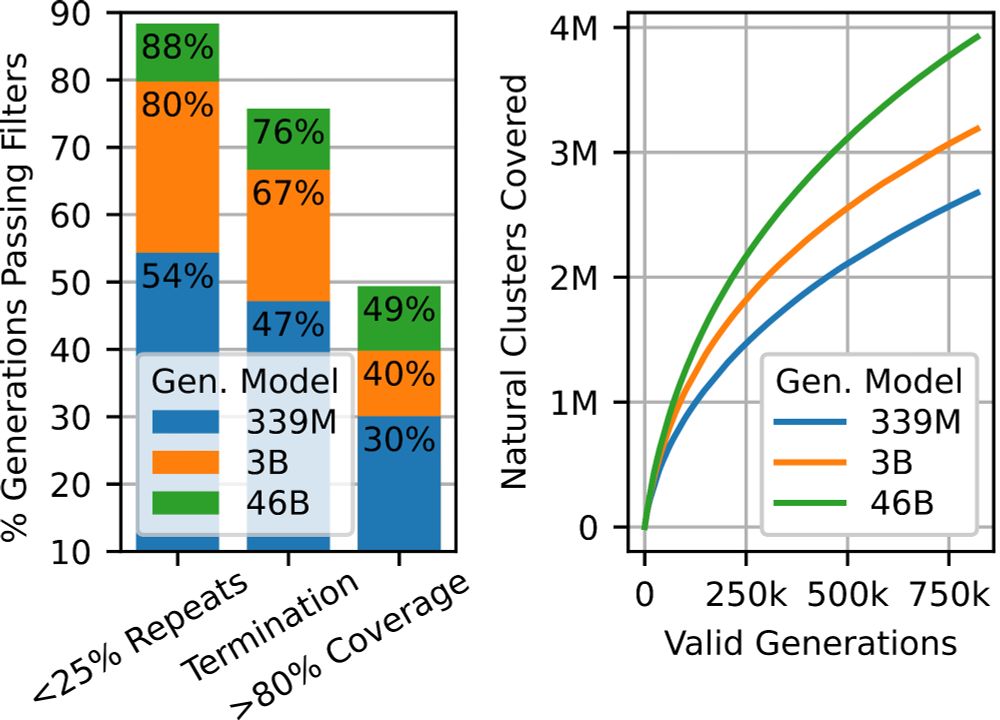
So what should we be evaluating? Generative models like ProGen3 are fundamentally trained to generate proteins. So we just let the models generate! We found that as models scale, not only do they generate higher quality sequences, but also produce considerably more diversity.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0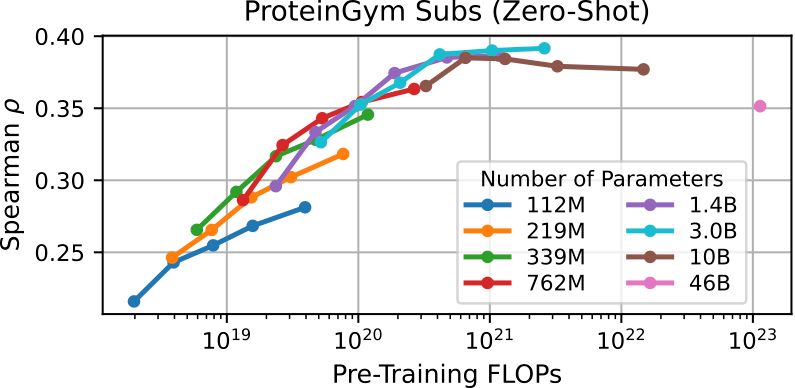
But why do all of this? What does scaling get us? ProteinGym is a nice benchmark for measuring zero-shot fitness prediction, but even three years ago (ProGen2) we found that this wasn’t the best proxy for evaluating scaling, and we still find that to be the case.
17.04.2025 20:28 — 👍 0 🔁 0 💬 1 📌 0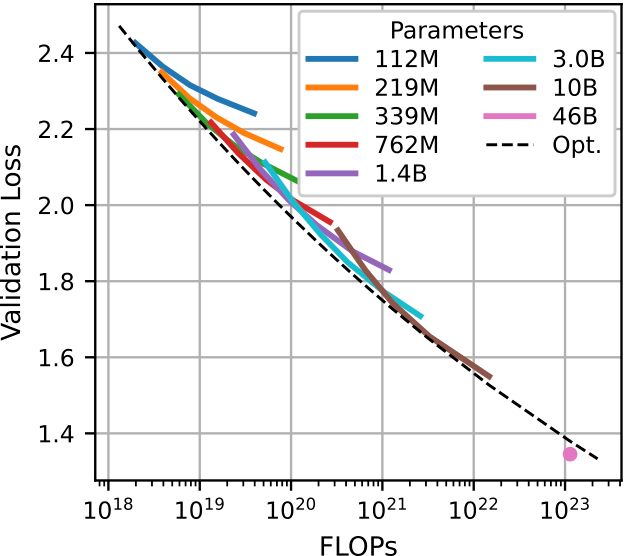
We developed optimal scaling laws that allowed us to scale up to 46B parameters, where we continue to see signs of generalization on diverse proteins far from the training data.
17.04.2025 20:28 — 👍 1 🔁 0 💬 1 📌 0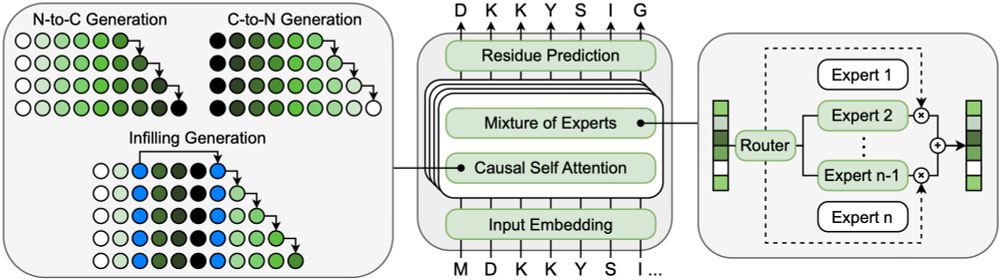
ProGen3 is a family of MoE models ranging from 112M to 46B parameters, capable of full sequence generation, as well as infilling. For practical protein design problems, having these new capabilities opens up a lot of new possibilities.
17.04.2025 20:28 — 👍 2 🔁 0 💬 1 📌 0
What does pushing the boundaries of model capacity and data scale do for generative protein language models? I’m super excited to share our latest work at Profluent Bio where we begin to explore and test some of our hypotheses!
www.biorxiv.org/content/10.1...
I’ll be in Vancouver for NeurIPS December 13-16, reach out if you’re interested in protein language models, genome editor / antibody design, or any of the other cool stuff we’re doing at Profluent!
06.12.2024 21:08 — 👍 5 🔁 0 💬 0 📌 0We note the 86% identity to S. cristatus Cas9 in the paper, but importantly found that OpenCRISPR-1 was <80% identity to anything in our database of patented sequences.
26.04.2024 01:28 — 👍 2 🔁 0 💬 1 📌 0Hey, happy to provide more info here! We did find other generated sequences with activity further from naturals (including S. cristatus), but for OpenCRISPR-1 we wanted a sequence that’d be viable as a drop-in for SpCas9 (high activity, NGG PAM), which ended up bringing us closer to a few naturals.
26.04.2024 01:27 — 👍 2 🔁 0 💬 1 📌 0