Check out what's next for 10x Genomics across #singlecell, #spatialbiology, and software!
19.02.2025 19:23 — 👍 4 🔁 0 💬 0 📌 0Dylan Webster
@dylanwebster.bsky.social
Software product at 10x Genomics. 🧬⚾🥋🎸
@dylanwebster.bsky.social
Software product at 10x Genomics. 🧬⚾🥋🎸
Check out what's next for 10x Genomics across #singlecell, #spatialbiology, and software!
19.02.2025 19:23 — 👍 4 🔁 0 💬 0 📌 0#singlecell has come a long way. But we're also just getting started!
06.02.2025 17:08 — 👍 0 🔁 0 💬 0 📌 0👀🤯
06.02.2025 17:06 — 👍 0 🔁 0 💬 0 📌 0👀 Spotted in the wild! The official 10x Genomics account has landed on Bluesky.
07.01.2025 22:14 — 👍 3 🔁 0 💬 0 📌 0Want to help build software that enables researchers to answer biology’s hardest and most elusive questions? Do so as Senior Product Manager, Cloud Visualization Software at
10x Genomics!
boards.greenhouse.io/10xgenomics/...
#singlecell #spatialbiology #hiring #productmanagement #TechJobs
Kudos to the Xenium Explorer team at 10x Genomics for consistently upleveling what's possible out of the box for Xenium analysis!
Check out our analysis page to see all Xenium analysis improvements! bit.ly/4flCNSu
#spatialbiology #insitu #singlecellspatial #bioinformatics
Want to help build software that enables researchers to answer biology’s hardest and most elusive questions? Do so as Senior Product Manager, Cloud Visualization Software at
10x Genomics!
boards.greenhouse.io/10xgenomics/...
#singlecell #spatialbiology #hiring #productmanagement #TechJobs

Emilie Schario says today's data tooling and LLMs make it so everyone can be a "data hire" 📊
26.11.2024 19:43 — 👍 1 🔁 1 💬 1 📌 0
Really good blog post!
"... [T]he answer isn’t SQL expertise; it’s curiosity... Remember, a data hire isn't just a job title; it's a mindset."
mixpanel.com/blog/emilie-...

Check out our new tools for #SpatialTranscriptomics. It's called Sainsc: onlinelibrary.wiley.com/doi/10.1002/....
#Sainsc can map cell types in high resolution spatial transcriptomics data, without segmenting cells! It's light weight, fast, and easy to use: sainsc.readthedocs.io/tutorials/in...
Wow!
26.11.2024 22:51 — 👍 1 🔁 0 💬 0 📌 0Extremely cool application of the Flex chemistry!
22.11.2024 18:45 — 👍 0 🔁 0 💬 0 📌 0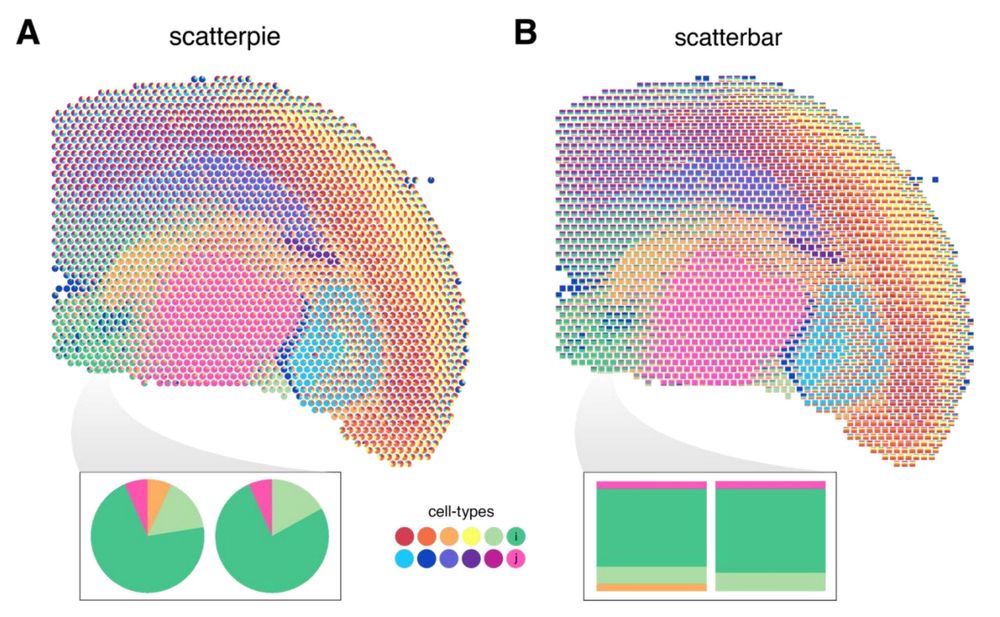
Check out our updated scatterbar package for visualizing proportional data across spatial coordinates like deconvolved cell types in #spatialtranscriptomics using stacked bar plots (alternative to scatterpies)!
Preprint: www.biorxiv.org/content/10.1...
Code: github.com/JEFworks-Lab...
#dataviz
And now you can try the automated cell annotation directly in Cell Ranger 9!
www.10xgenomics.com/support/soft...
#singlecell #bioinformatics #genomics
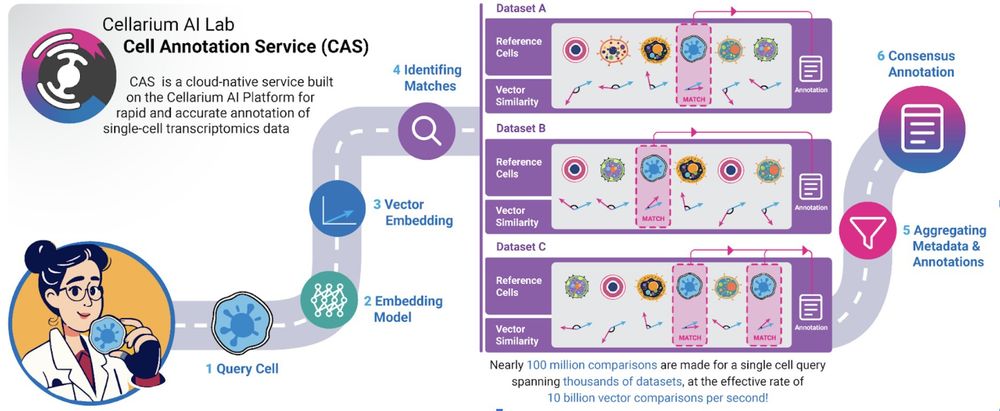
Infographic of the Cell Annotation Service
Super excited about this cell typing collab between 10x Genomics and the Broad. broad.io/CAS-News
Try it for free on cloud.10xgenomics.com/cloud-analysis and look out for more in the next Cell Ranger release coming imminently ;) (Let us know what you think!)
#singlecell #bioinformatics #genomics
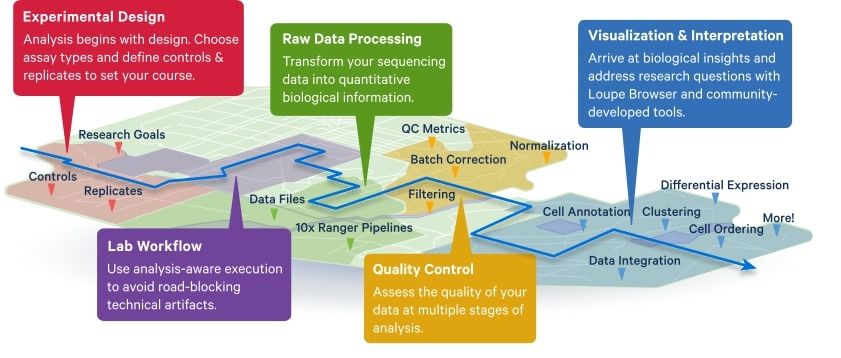
Overview of the roadmap for 10x Genomics data analysis
Did you know that 10x Genomics publishes intros, tutorials, and blogs that help navigate the journey to publish #SingleCell, Visium #SpatialTranscriptomics and Xenium #singlecellspatial data? Check out the Analysis Guides!
www.10xgenomics.com/resources/an...
#Genomics #Bioinformatics
You can also check out the public datasets that show this plexy: www.10xgenomics.com/resources/da...
04.10.2023 04:34 — 👍 1 🔁 0 💬 0 📌 0There are 18,536 genes in the human probe set and 20,551 in the mouse probe set.
This page also documents the Human Immune Cell Profiling Panel Antibody List for simultaneous readout of 35 proteins on Visium: www.10xgenomics.com/support/spat...
Hi Albert, this document has an overview of both the poly(A) capture approach for FF and the probe based approach for FFPE: www.10xgenomics.com/resources/do...
This page has detailed documentation of the FFPE probe sets (click through to the individual pages: www.10xgenomics.com/support/spat...

Download the new Loupe here:
www.10xgenomics.com/support/soft...
Our goal is to accelerate your journey to experimental insights, and we hope you give Loupe 7 & and LoupeR a spin to see for yourself!
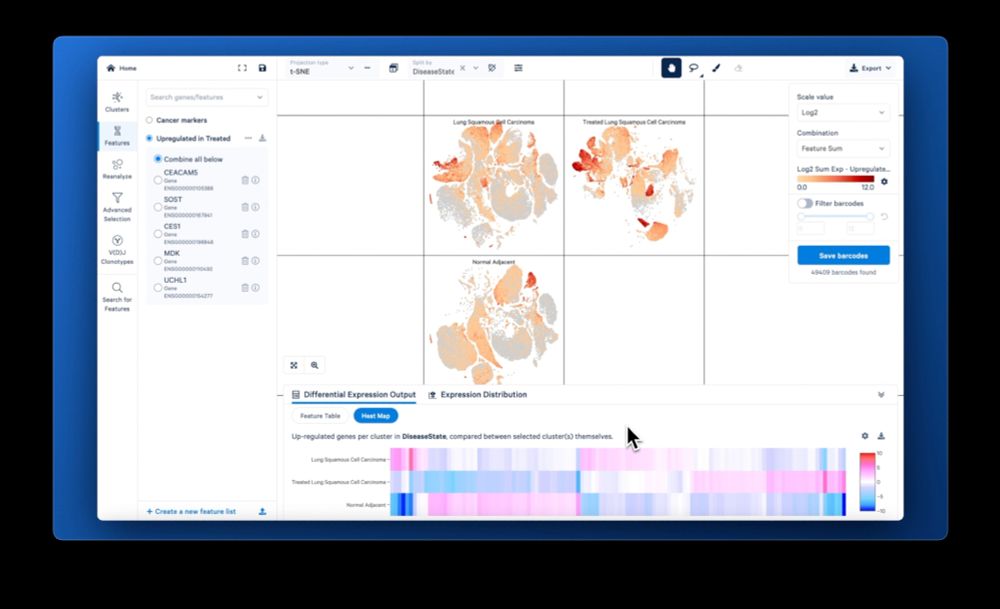
And check out how the redesigned Loupe user interface makes it simple to visualize the expression of your favorite genes!
02.10.2023 15:38 — 👍 0 🔁 0 💬 1 📌 0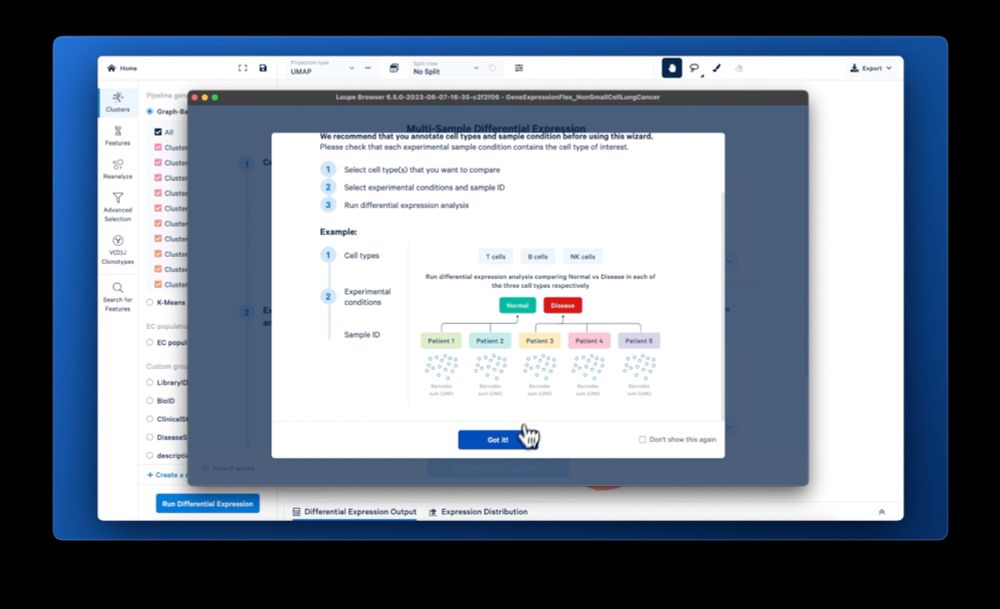
The feature I’m most excited about: Finding target genes in your experiment just got easier with the new multi-sample differential expression in Loupe 7
02.10.2023 15:38 — 👍 0 🔁 0 💬 1 📌 0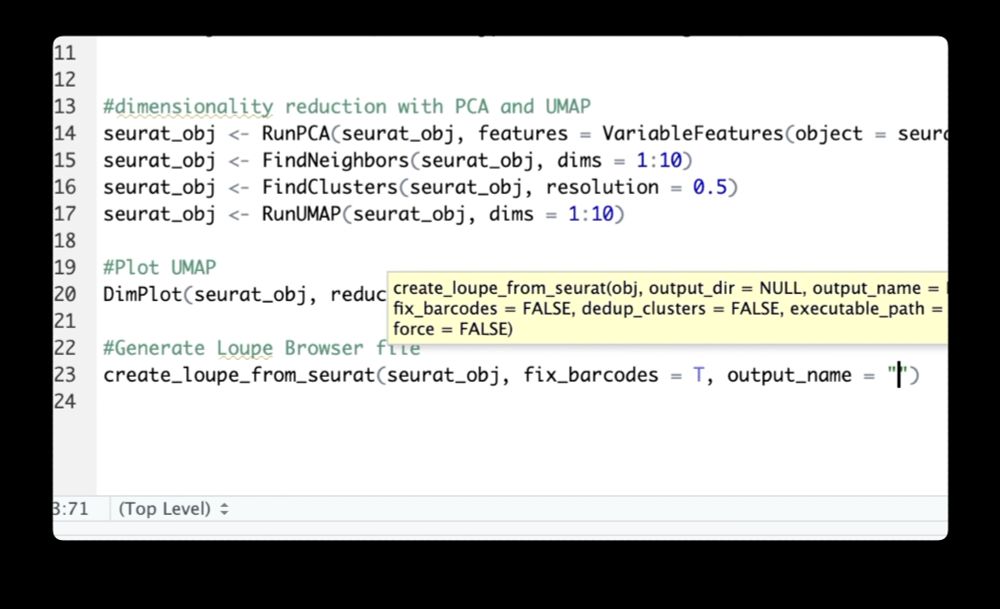
Want to analyze Seurat data without code? Use the new 10x Genomics LoupeR package to move your data into the new Loupe 7. It sports a redesigned UI, and includes new features that put publication-ready figures just a few clicks away!
#genomics #singlecell
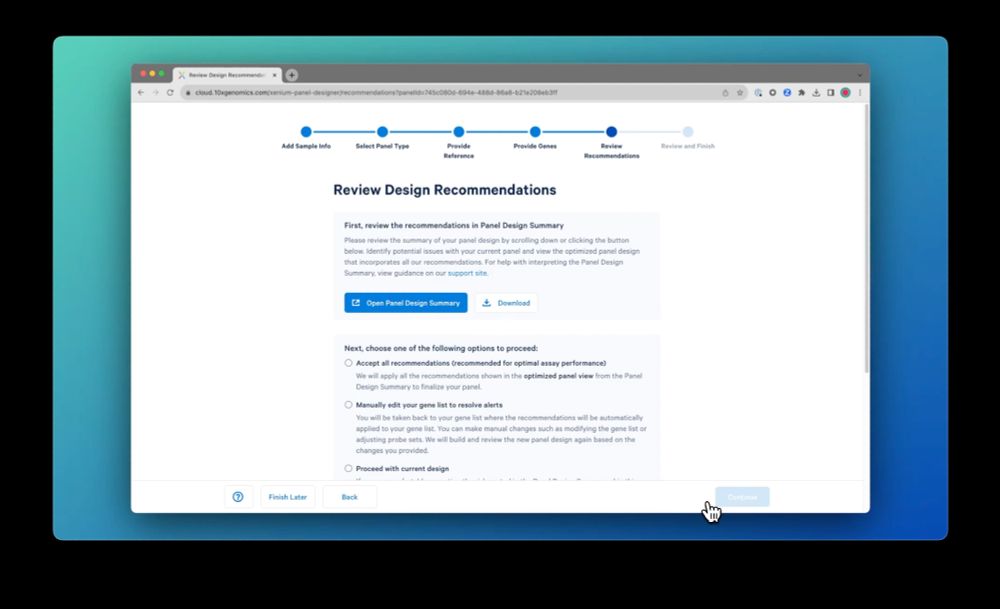
Finally, review the recommendations for optimal assay performance. Either accept the recommendations or iterate as needed. At the end of the workflow, your panel is ready to be ordered and manufactured. Check it out for yourself!
cloud.10xgenomics.com/xenium-panel...
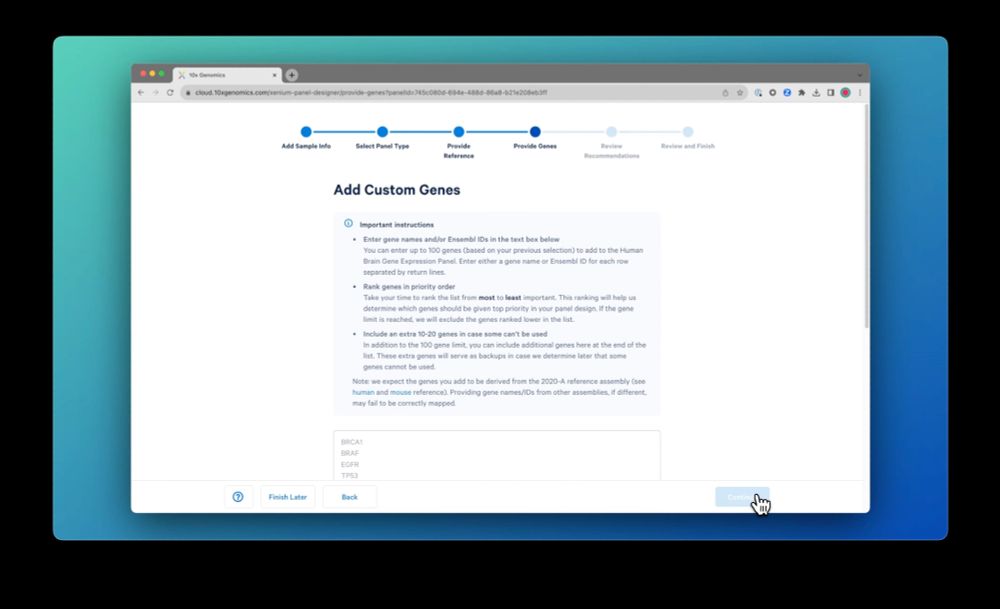
Next, paste the list of genes you want to include. You can easily clean up the list to resolve any ambiguous gene names, etc. Then start building the design when you’re done.
02.10.2023 15:31 — 👍 0 🔁 0 💬 1 📌 0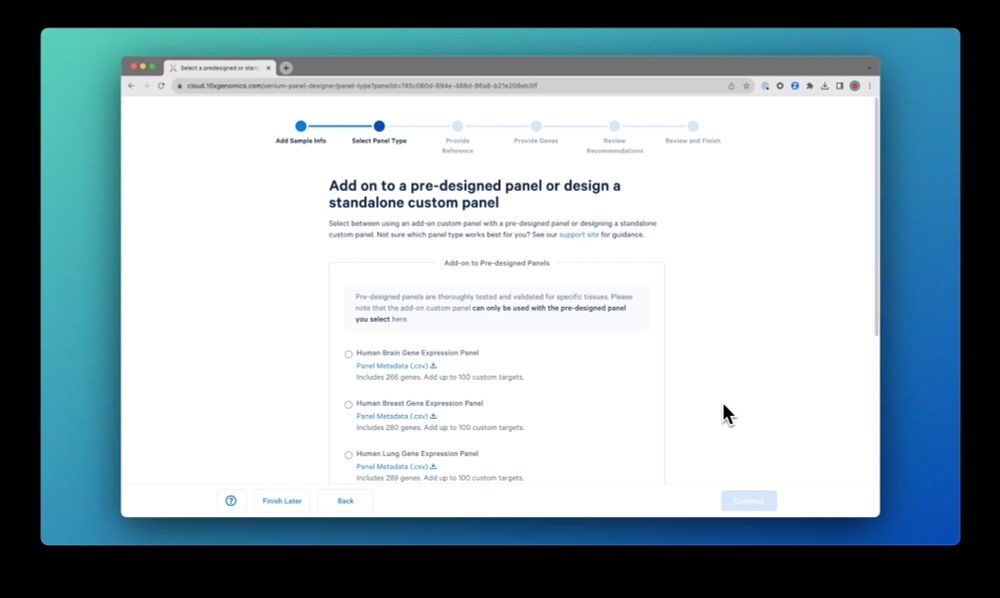
Easily design 10x Genomics #singlecellspatial custom panels with the new #Xenium Panel Designer: Start with a pre-designed panel, or build a standalone custom panel (up to 480 plex). Design against curated references from CELLxGENE and GEO or bring your own references.
02.10.2023 15:31 — 👍 0 🔁 0 💬 1 📌 0
Read all about how Xenium analysis shortens the path to insight at www.10xgenomics.com/products/xen...
02.10.2023 15:27 — 👍 0 🔁 0 💬 0 📌 0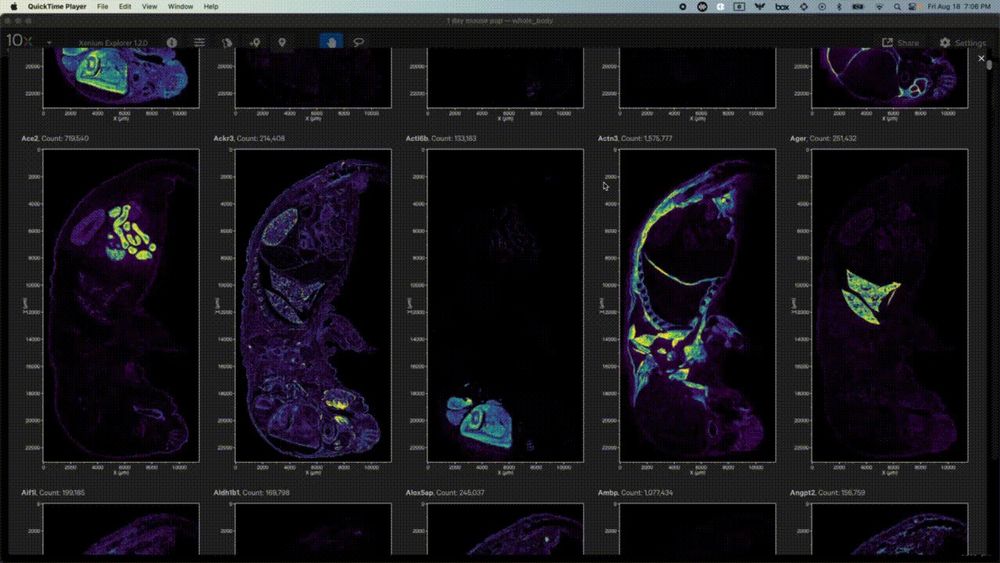
Use the per-gene localization gallery to fly through expression of each individual gene.
02.10.2023 15:24 — 👍 0 🔁 0 💬 1 📌 0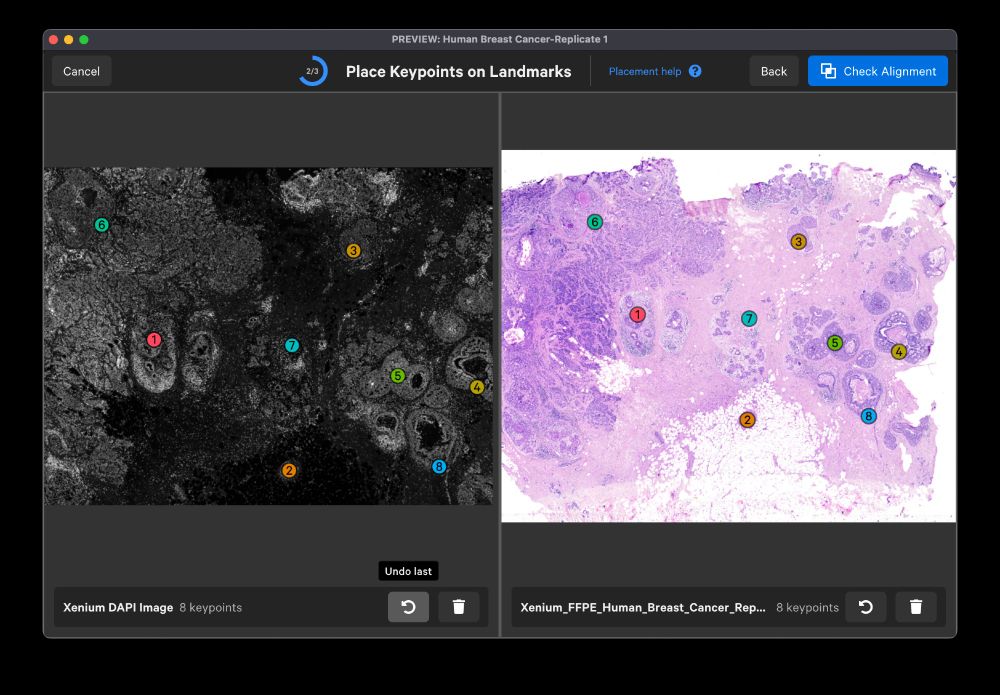
If you do H&E on the same section as your Xenium run, XE 1.2 also provides an intuitive workflow to align and overlay H&E in the Xenium coordinate space so you can see sub-cellular gene expression on top of your histopathological ground truth.
02.10.2023 15:23 — 👍 1 🔁 0 💬 0 📌 0The Xenium Explorer screenshot represents exactly what can be visualized directly following a Xenium instrument run—where transcripts are decoded, cells are segmented, and graph-based clustering is computed as soon as the run is done with Xenium Onboard Analysis.
02.10.2023 15:23 — 👍 0 🔁 0 💬 2 📌 0