I think missing annotations is the problem here. Until we have more complete sequenced proteomes, global phylogeny is likely to shift in the future
08.02.2026 09:38 — 👍 1 🔁 0 💬 0 📌 0
Huge congrats to lead authors Qiuzhen Li & Diandra Daumiller! Thanks to @martinsteinegger.bsky.social for all the great tools!
📄 Preprint: biorxiv.org/content/10.6...
🖥️ Try the Tool: she-app.serve.scilifelab.se
07.02.2026 08:50 — 👍 9 🔁 0 💬 0 📌 0

🕊️ Structural Acceleration
Does a bigger genome mean faster structural evolution? No.
We found lineage-specific bursts of structural innovation in Birds (Aves) and Ants (Hymenoptera) that are distinct from genomic expansion. (4/5)
07.02.2026 08:50 — 👍 4 🔁 0 💬 1 📌 0
⚙️ A Bipartite Evolutionary Mode
The eukaryotic proteome isn't a uniform soup.
We resolve it into two distinct modes:
A rigid "Architectural Core" (Cytoskeleton/Chaperones)
A highly plastic "Operational Engine" (Metabolism/Translation) (3/5)
07.02.2026 08:50 — 👍 5 🔁 1 💬 1 📌 0

🐁 Stop guessing your model organism.
Our new search engine allows you to rank model organisms by their structural fidelity to specific human pathways.
Case Study: For Fanconi Anaemia, Mouse is perfect. Yeast fails. C. elegans is a hidden gem. (2/5)
07.02.2026 08:50 — 👍 9 🔁 0 💬 1 📌 0

Introducing The Structural History of Eukarya (SHE): The first proteome-scale phylogeny constructed entirely from 3D structure.
We computed 300 trillion alignments across 1,542 species to map the tree of life. 🧵👇 (1/5)
07.02.2026 08:50 — 👍 84 🔁 40 💬 2 📌 0

The future of drug design is in AI. RareFoldGPCR: Agonist Design Beyond Natural Amino Acids.
Paper: www.biorxiv.org/content/10.1...
Code: github.com/patrickbryan...
03.10.2025 15:09 — 👍 2 🔁 0 💬 0 📌 0
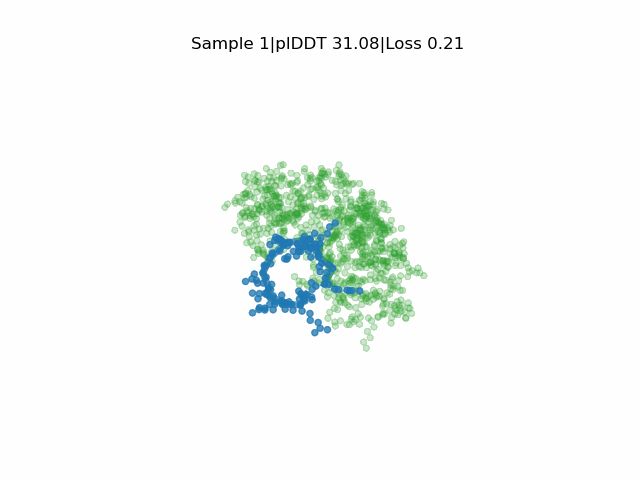
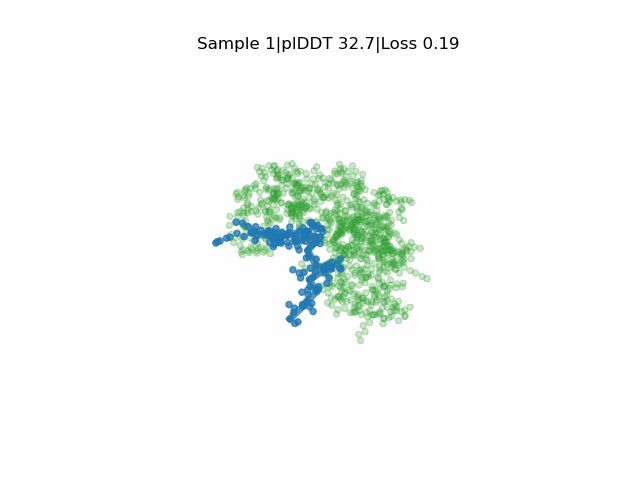
Our study where we develop EvoBind2: Design of linear and cyclic peptide binders from protein sequence information is now published! www.nature.com/articles/s42...
22.07.2025 11:23 — 👍 4 🔁 2 💬 1 📌 0
Cool! Congrats @proteinator.bsky.social 🎉
17.07.2025 05:49 — 👍 2 🔁 0 💬 0 📌 0
We have much more coming in this space where we can identify target interfaces and inhibit the interactions - all using protein structure prediction!
06.07.2025 08:02 — 👍 0 🔁 0 💬 0 📌 0
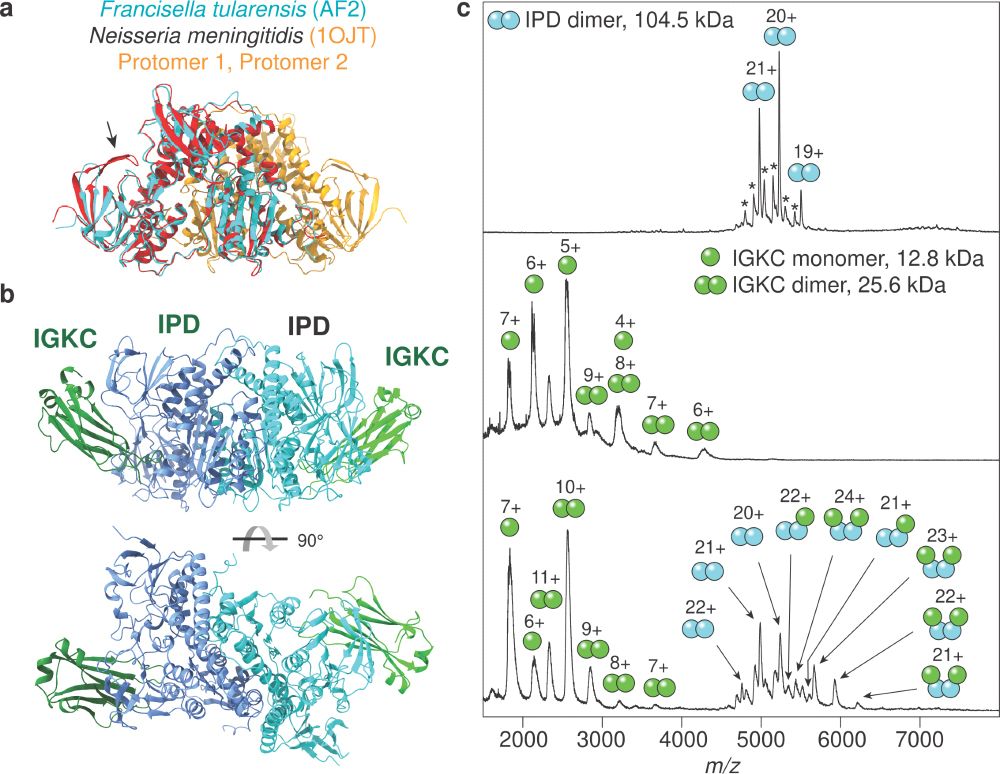
Now published: our study on human-pathogen protein-protein interactions! We identify 30 interactions with an expected TM-score ≥0.9, tripling the structural coverage in these networks. One novel interaction was validated with mass spectrometry. journals.plos.org/ploscompbiol...
06.07.2025 08:02 — 👍 4 🔁 0 💬 2 📌 0
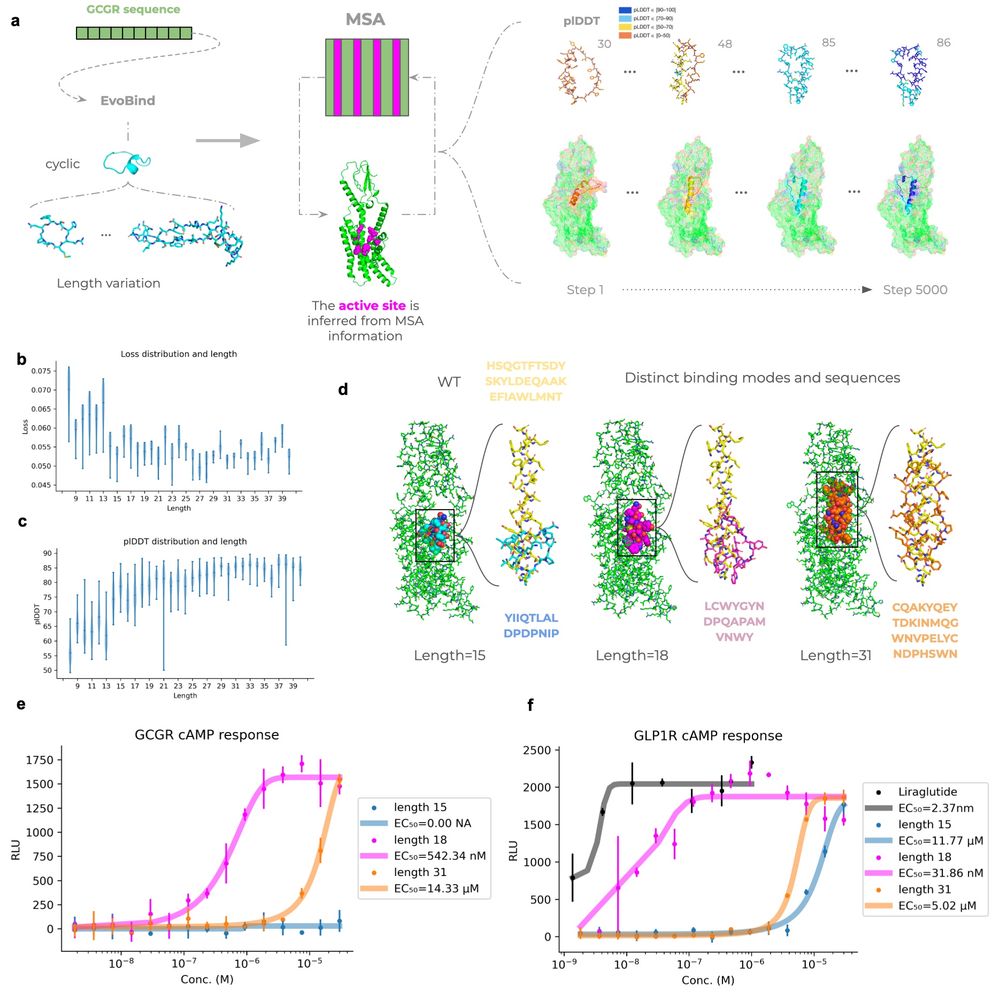
Our latest work is out: we designed dual GLP1R/GCGR agonists—cyclic peptides that activate both metabolic receptors, entirely from sequence alone.
This has never been done before. www.biorxiv.org/content/10.1...
09.06.2025 07:00 — 👍 5 🔁 1 💬 0 📌 0
You can also design in Colab now: colab.research.google.com/github/patri...
27.05.2025 12:40 — 👍 8 🔁 2 💬 0 📌 0
The WT binder is 1.8 uM which means that we create as good binders but with new modes of binding for a target where these NCAA interactions are completely unknown 😎
25.05.2025 20:04 — 👍 1 🔁 0 💬 0 📌 0
Thanks! We will release a lot of new tech this year - stay tuned! We are only in the beginning of protein design I think
25.05.2025 20:02 — 👍 2 🔁 0 💬 0 📌 0
Just like we have used EvoBind to e.g. create functional HIV inhibitors in a single shot (biorxiv.org/content/10.1...) we can now do this with an expanded vocabulary to have more chemical possibilities and avoid e.g. immune recognition and degradation
25.05.2025 14:11 — 👍 0 🔁 0 💬 1 📌 0
RareFold supports 49 different AAs.
The 20 regular, and 29 rare ones: MSE, TPO, MLY, CME, PTR, SEP,SAH, CSO, PCA, KCX, CAS, CSD, MLZ, OCS, ALY, CSS, CSX, HIC, HYP, YCM, YOF, M3L, PFF, CGU,FTR, LLP, CAF, CMH, MHO.
You can simply specify which you want to use and design!
25.05.2025 14:11 — 👍 0 🔁 0 💬 2 📌 0
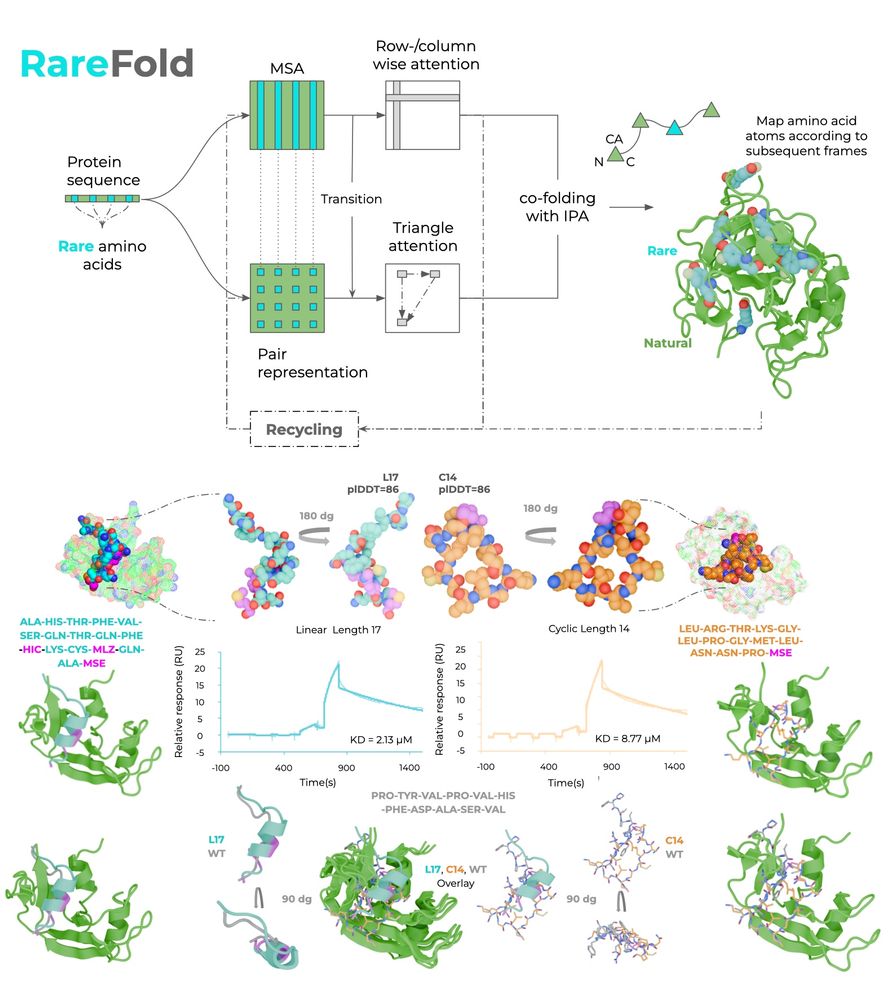
Happy to release our breakthrough AI-model: RareFold, which predicts and designs proteins with noncanonical AAs. With EvoBindRare, we designed linear & cyclic peptide binders with high affinity & novel binding modes, wet lab validated.
📄 biorxiv.org/content/10.1...
💻 github.com/patrickbryan...
25.05.2025 14:11 — 👍 19 🔁 4 💬 2 📌 1
RNA biology lab studying cell fate and cell identity in the context of embryonic development and disease @Goethe University
#ribosome #translational_control #RBP #splicing #RNA_localizatilzon
https://kurianlab.com/
Alum:@salkinstitute @yeo lab, UCSD
Predoctoral Research Assistant at the Llorca lab @cniostopcancer.bsky.social working on mTORC1 signaling and cryo-EM ❄️🔬. Intrigued by the structural mechanisms of protein assemblies involved in endomembrane signaling and trafficking 🧪.
Computational chemist/structural bioinformatician working on improving molecular simulation at MRC Laboratory of Molecular Biology. jgreener64.github.io
Developing data intensive computational methods • PI @ Seoul National University 🇰🇷 • #FirstGen • he/him • Hauptschüler
PhD candidate in bioinformatics
Protein structure prediction / Protein design
Structural biology and protein engineering
Asst. Prof. Uni Groningen 🇳🇱
Comp & Exp Biochemist, Protein Engineer, 'Would-be designer' (F. Arnold) | SynBio | HT Screens & Selections | Nucleic Acid Enzymes | Biocatalysis | Rstats & Datavis
https://www.fuerstlab.com
https://orcid.org/0000-0001-7720-9
PhD Fellow @cphbiosciphd @NMITaylorLab @UCPH_CPR 🇩🇰 | Structural biology 🤝 phages 🔬 🇧🇷 (he/him)
Assistant Professor at UMich with a focus on computational pharmacology (he/him)
Computational physicist at https://peptone.io
PhD @GroupParrinello, PostDoc @franknoe.bsky.social
Disordered Proteins, AI for Science, Molecular Dynamics, Enhanced Sampling
🔗 https://scholar.google.com/citations?user=fnJktPAAAAAJ
Postdoc in the Pauli Group at IMP Vienna studying fertilization | #scicomm | #mentoring | #scienceandfaith | #DiversityandInclusion | Salvadoran scientist 🇸🇻
Biologist that navigate in the oceans of diversity through space-time
Protein evolution, metagenomics, AI/ML/DL
Website https://miangoaren.github.io/
Associate Professor @ University of Sao Paulo. Structural biologist. Protein crystallographer. Interested in protein-ligand interactions and modeling
PhD Student @TU_Muenchen Previous @TheDPTechnology | @chalmersuniv | @VolvoGroup | BSc. NUDT 18'
https://weilong-web.github.io
Postdoctoral researcher in computational structural biology at the University of Copenhagen with Kresten Lindorff-Larsen
Software Engineer, Writer, curious about distributed social media applications
Bio x ML @ Romero lab, @dukeubme.bsky.social
@ml4proteins.bsky.social co-organizer