There should be support for ECRs as well. There are also country specific programs, I know Norway, Sweden and Denmark have recently announced their own programs. Just type in country of interest and keep checking, I assume most of EU countries will announce similar programs this year.
05.05.2025 23:31 —
👍 1
🔁 0
💬 0
📌 0
EU just pledged €0.5B in research funding to attract and retain talent👇 The initiative is called 'Choose Europe for Science'
05.05.2025 23:18 —
👍 4
🔁 2
💬 1
📌 0

Thank you for joining us at US HUPO 2025 in Philadelphia! We will see you in St. Louis for US HUPO 2026! #Proteomics #USHUPO2025 #USHUPOConference 🧪
26.02.2025 18:30 —
👍 17
🔁 5
💬 2
📌 0
How good or bad idea is it to take one week vacation and be dropped the evening before #USHUPO2025 starts (and you lead morning short course 🚀)? Guess we will find out tomorrow. Ping me here or on LinkedIn if you want to chat. I am happy to find time! #Proteomics
23.02.2025 02:47 —
👍 3
🔁 0
💬 1
📌 0
🤔 Ever wondered how proteomics thrives in (bio)pharma🚀? How you could provide impact in industry with proteomics background earned in academia?
Please join me and @drawheatcraft.bsky.social for Sunday morning US HUPO 2025 short course 👇
15.01.2025 19:44 —
👍 11
🔁 7
💬 1
📌 0
We will look at proteomics in pharma industry from multiple angles
🌐How it integrates into the pharma matrix environment.
🎯How it brings impact and propels our knowledge.
🤝How to learn more about proteomics in industry through various forms of employment or collaborations.
15.01.2025 19:45 —
👍 0
🔁 0
💬 0
📌 0
🤔 Ever wondered how proteomics thrives in (bio)pharma🚀? How you could provide impact in industry with proteomics background earned in academia?
Please join me and @drawheatcraft.bsky.social for Sunday morning US HUPO 2025 short course 👇
15.01.2025 19:44 —
👍 11
🔁 7
💬 1
📌 0
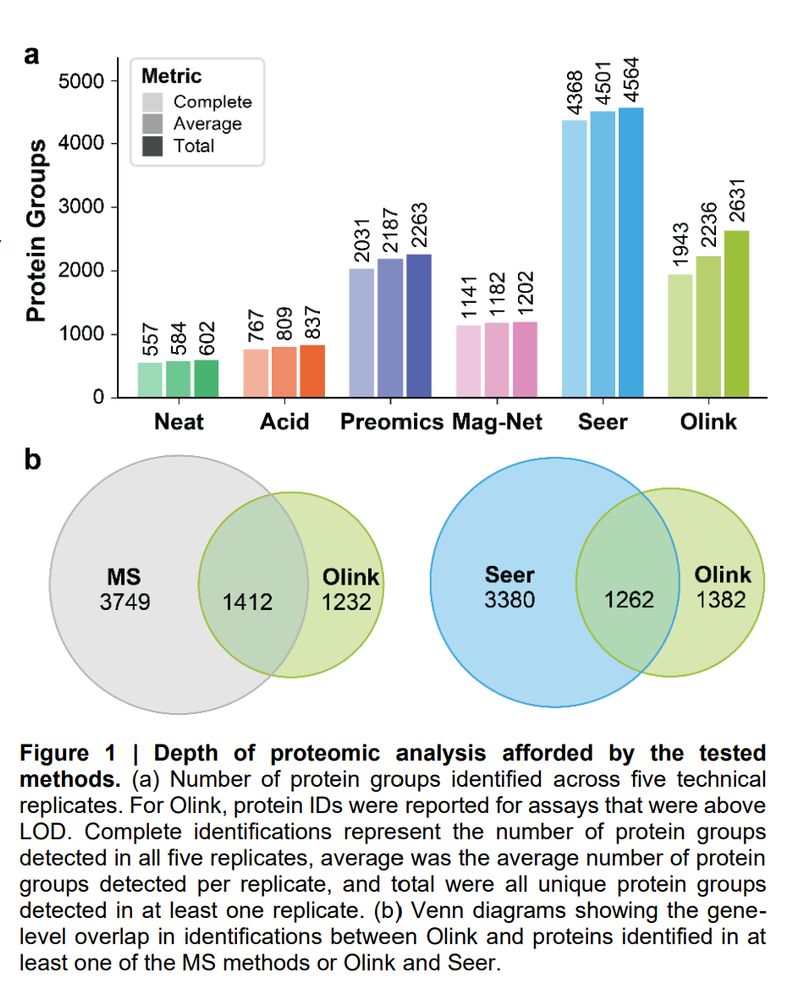
A Technical Evaluation of Plasma Proteomics Technologies www.biorxiv.org/cont...
---
#proteomics #prot-preprint
13.01.2025 16:20 —
👍 19
🔁 4
💬 3
📌 1
May Institute – Computation and statistics for mass spectrometry and proteomics
May Institute on Computation and Statistics for Mass Spectrometry and Proteomics @KhouryCollege @Northeastern in Boston MA is happening in person on April 28-May 11, 2025 and is accepting applications! You will not regret attending computationalproteomics.khoury.northeastern.edu
12.01.2025 16:50 —
👍 50
🔁 28
💬 1
📌 1
May Institute – Computation and statistics for mass spectrometry and proteomics
The May Institute is a fantastic learning experience and available virtually and in person! #teammassspec #proteomics computationalproteomics.khoury.northeastern.edu
12.01.2025 16:29 —
👍 6
🔁 2
💬 0
📌 0
...Looks like I have to force myself finally write that paper on this 😅
12.01.2025 00:09 —
👍 2
🔁 0
💬 1
📌 0
I also just remembered there was another, scientific, reason. When enriching protein the molar binding capacity is actually lower than when enriching peptides - some binging sites will become inaccessible because they are spatially blocked by the protein size. This was advantageous in our settings.
12.01.2025 00:05 —
👍 2
🔁 0
💬 1
📌 0
Economic reasons 🙂 the biotynated proteins make up only ~2% of the sample. Digesting enriched fraction resulted in 50fold savings in trypsin cost I think we went from $25k to $500 for one study.
11.01.2025 23:52 —
👍 2
🔁 0
💬 1
📌 0
Happy to help🙂
11.01.2025 14:49 —
👍 1
🔁 0
💬 0
📌 0
If those look good and issues prevail, I fall back to some relatively accessible positive control samples to pinpoint the issue. Alternatively you can use antibody against biotin for pull down to bypass streptavidin and check that the sample quality is good. Good luck.
11.01.2025 02:53 —
👍 2
🔁 0
💬 1
📌 0
That's too difficult to say. I would try with fresh batch of beads and if that doesn't help look for other reasons for failure. IDs are the ultimate read out, it is important to have reliable check points along the way like the yield and protein concentration if the import sample.
11.01.2025 02:51 —
👍 1
🔁 0
💬 1
📌 0
I discard the non-biotinglated peptides. Too high likelihood that some come from non-specific binding. Unless you need to pool to get over the sensitivity threshold. Even in that case I would only consider hits where there is biotin tag/specific fragment detected on given peptide/protein.
11.01.2025 01:57 —
👍 2
🔁 0
💬 2
📌 0
Absolutely, just want to mention than in order to optimize the labeling steps it it essential to understand the performance of the pull down and it's reliability (hence the need for positive control like the biotinylated cell lysate or similar).
11.01.2025 01:18 —
👍 2
🔁 0
💬 0
📌 0
And for elution. We also did on-bead digestion which allows for elution with high acid+ACN steps -> dry down resuspend in LC-MS injection buffer. There are some publications on this. If you tune all steps you can get to >90% peptides biotinylated in the final sample.
11.01.2025 00:49 —
👍 1
🔁 0
💬 1
📌 0
Can be hard. I would make a rough estimate from the prevalence of the PTM. We have done similar thing where we specifically targeted small % of proteome. A good positive control sample was biotynating HeLa lysate and then mixing it unlabeled HeLa in expected ratios. Allows MD without precious sample
10.01.2025 21:38 —
👍 1
🔁 0
💬 0
📌 0
Also, I've heard there are some bad batches of streptavidin going around.
10.01.2025 20:54 —
👍 1
🔁 0
💬 1
📌 0
For us the main optimization factor was carefully matching binding capacity to expected amount of biotinylated protein. Or rather making sure there is more biotinylated protein in the sample then binding capacity. The unspecific binding is basically unwashable.
10.01.2025 20:47 —
👍 2
🔁 0
💬 2
📌 0
How come this beautiful preprint didn't get much traction in here🤔? We will definitely want to give MAETi a try! Anyone else is planning to try this out? Any words of wisdom from team #proteomics?
04.01.2025 12:31 —
👍 5
🔁 0
💬 1
📌 0
Honestly don't know. I found it in my Google Chrome 'news' feed😁 and since my brain is on vacation this time of the year too I didn't search the source... It looks like it is AI generated based on the YT channel info.
29.12.2024 13:43 —
👍 1
🔁 0
💬 0
📌 0
YouTube video by 2 Minute Expert
Proteomics in 2 Minutes
I am a proteomicist 🧑🔬 nice to meet you 👋 #teammassspec #proteomics youtu.be/b3ImaM7v2OY?...
28.12.2024 02:28 —
👍 25
🔁 4
💬 3
📌 1
UPDATE: position filled! Thanks everyone for the help🙂
20.12.2024 11:15 —
👍 2
🔁 0
💬 0
📌 0
We Are the Analyzers - A Proteomics Anthem by US HUPO.
We Are the Analyzers - Proteomics Anthem
We’re thrilled to present "We Are the Analyzers" – an anthem for the proteomics community! A huge shoutout to everyone who brought this incredible song to life. Look what proteomics has already shown us 🎶🧪youtu.be/wzmJDNmsWK8 #Proteomics #USHUPO20YRS
18.12.2024 19:00 —
👍 22
🔁 12
💬 1
📌 4






