
We are happy to share that the preprint for Proteobench is available now on biorxiv!
Check it out here: www.biorxiv.org/content/10.6...
Congratulations and thanks to everyone involved for the great effort!
@drmuth.bsky.social
Group Leader and Adjunct Professor 🧑🏫 | Public Health | Data Engineering & Visualization | 🧬 Bioinformatics & 📈 Mass Spec Enthusiast | 📍 Berlin | Passionate About Collaboration and Open Science 🔬✨

We are happy to share that the preprint for Proteobench is available now on biorxiv!
Check it out here: www.biorxiv.org/content/10.6...
Congratulations and thanks to everyone involved for the great effort!

Preprint: Accurate de novo sequencing of the modified proteome with OmniNovo
arxiv.org/abs/2512.12272

New preprint! 🚀
We introduce AbTags – antibody–peptide fusion proteins that massively amplify signal, enable absolute quantification of binding events, and are read out together with the single-cell proteome by LC–MS.
AbTag platform is patent-pending.
Preprint: www.biorxiv.org/content/10.6...

Super excited to have this paper out. We developed a complex set of ground truth samples to test more than 110 statistical approaches for differential #metaproteomics
We can now give clear guidance on what tests perform well. Thank you Tjorven Hinzke and
@benoitkunath.bsky.social for the leadership

Meta-PepView: a metaproteomics performance evaluation and visualization platform
www.biorxiv.org/content/10.6...

Preprint: Reflection Pretraining Enables Token-Level Self-Correction in Biological Sequence Models
arxiv.org/abs/2512.20954

Modanovo: A Unified Model for Post-Translational Modification-Aware de Novo Sequencing Using Experimental Spectra from In Vivo and Synthetic Peptides - Molecular & Cellular Proteomics www.mcponline.org/article/S153...
25.12.2025 20:13 — 👍 3 🔁 1 💬 0 📌 0
The solution is right above us. Solar panels on existing buildings, car parks, and along highways generate clean power without sacrificing farmland or wild spaces. Let's use our built world first. #Renewables #Innovation
22.12.2025 02:27 — 👍 3879 🔁 932 💬 212 📌 71
HDSE-MS: Tandem Mass Spectrum Prediction for Small Molecules via Hierarchical Distance Structural Encoding | Analytical Chemistry pubs.acs.org/doi/10.1021/...
10.12.2025 19:48 — 👍 0 🔁 0 💬 0 📌 0
Researchers have performed a large-scale genetic screen to uncover the hidden roles of #microproteins. One of the discovered microproteins, named PIPPI, was found to protect cells from stress in the endoplasmic reticulum. Published in @narjournal.bsky.social 🧪 #proteomics news.ki.se/new-micropro...
27.11.2025 07:55 — 👍 8 🔁 4 💬 1 📌 0
Cannot hide my joy, relief, excitement being able to share the outcome of persevering (to say the least) collaborative work. So... the MS glyco(proteomics) guidelines are published!! www.mcponline.org/article/S153...
Many thanks to @beilstein-institut.bsky.social @glycosmos.bsky.social @sib.swiss

It’s the first time that proteomics has been used to analyze Renaissance recipes. The project demonstrates an innovative way to study medicine and the circulation and use of medical recipes from the past. cen.acs.org/analytical-c... #chemsky 🧪
27.11.2025 18:16 — 👍 15 🔁 7 💬 0 📌 2This project started 5 years ago. It led us to add isotope-labeling support to #FragPipe/#IonQuant. Since then, the tools have grown so much and are now widely used in #Chemoproteomics.
Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social

ProteomeXchange consortium in 2026: making proteomics data FAIR url: academic.oup.com/nar/article/...
09.11.2025 19:49 — 👍 2 🔁 0 💬 0 📌 0
On the event of James Watson's death, I highly recommend this 2023 commentary from @matthewcobb.bsky.social and Nathaniel Comfort with crucial new insights into the discovery of the double helix. (And also check out Cobb's brand new biography of Francis Crick) www.nature.com/articles/d41...
07.11.2025 21:25 — 👍 268 🔁 122 💬 7 📌 7
Screenshot of a Tweet pointing out that academic spam is actually in its own way kind of encouraging.
I think about this post every day 🧪
07.11.2025 12:15 — 👍 640 🔁 111 💬 3 📌 1The reaction to this is fascinating. It's like a microcosm of all discussions around AI: lots of enthusiasm, lots of loathing (much of it reflexive), and some wise 'let's maybe try it and see' responses.
07.11.2025 13:43 — 👍 35 🔁 12 💬 11 📌 4Finally, the R package we needed, but don't deserve. I can already hear @benneely.com cackling from across the country.
Thanks to @willfondrie.com for alerting me to this amazing repo.

This is figure 1, which shows abundance, transcription and distribution of hydrogenase genes and H2-related metabolic genes throughout the human gut.
A previously uncharacterized microbial enzyme is responsible for the production of molecular hydrogen in the gut, which drives the growth of other bacteria and has implications for human health, according to a paper in Nature Microbiology. go.nature.com/4oKOveG #microbiome #medsky 🧪
05.11.2025 14:08 — 👍 18 🔁 5 💬 1 📌 0
🧬 Thrilled to share our latest paper in @natmicrobiol.nature.com 📄
A collaboration to give the Flaviviridae (home to Zika, Dengue & HCV) a much-needed taxonomic re-think.
Our at-scale AI structure prediction gave a complementary perspective on viral evolution.
www.nature.com/articles/s41...
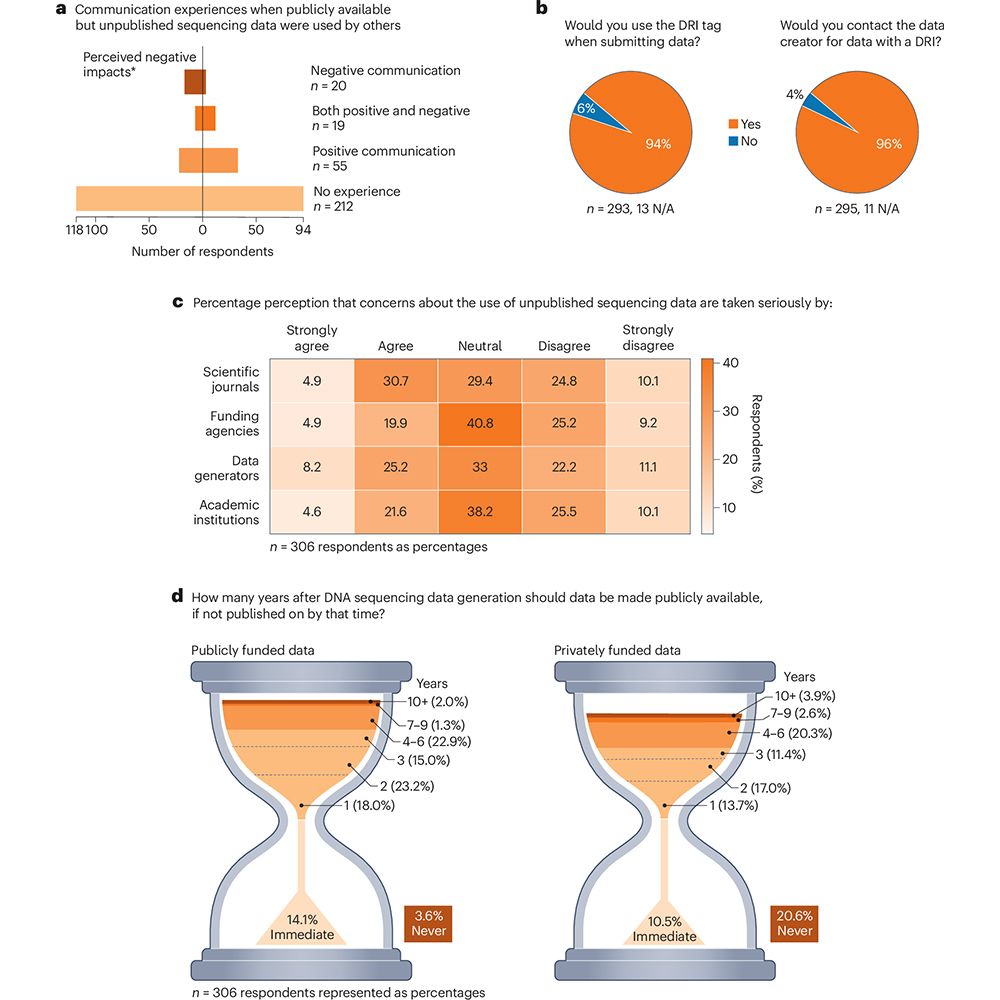
This is figure 1, which shows the summary results from a survey of 306 scientists on data reuse.
In a Consensus Statement in Nature Microbiology, a consortium of #microbiome scientists discusses current sequencing data sharing policies and proposes the use of a Data Reuse Information tag to promote equitable and collaborative data sharing. go.nature.com/4o1Gl1f 🧪
30.09.2025 13:15 — 👍 19 🔁 11 💬 1 📌 0
Our latest paper introduces new methods for improving peptide retention time predictions in proteomics, incorporating chemical structure information to better handle unseen modifications.
Read all about it in our preprint by👉 doi.org/10.1101/2025...
#Proteomics #AI #MachineLearning

Carafe - spectral library prediction for DIA proteomics
www.nature.com/articles/s41...
If you are interested in mRNA vaccines against bacteria, take a look at our recent review. It was a pleasure to write this together with all co-authors! rdcu.be/eyjBq
www.nature.com/articles/s41...
Sure, I know! I lived in Wedding for a while. 😉
01.11.2025 22:18 — 👍 1 🔁 0 💬 1 📌 0Would have been fun to realize that we are actually at the same spot-the same time… 😆
01.11.2025 20:48 — 👍 1 🔁 0 💬 2 📌 0
“We fundamentally believe that publishing less – but better – is essential for the health of the entire research system worldwide,” the authors of the report state.
21.10.2025 19:11 — 👍 90 🔁 32 💬 0 📌 9
I was there, the same day!!! Just checked the meta data of my photo. The almost only difference, it was dark… 😉
01.11.2025 20:37 — 👍 1 🔁 0 💬 1 📌 0
From Identification to Insight: Making Full Use of the Diagnostic Potential of MS/MS Proteotyping in Clinical Microbiology Using Efficient Bioinformatics pubs.acs.org/doi/10....
---
#proteomics #prot-paper