Trainees: If you haven't already, learn the value of implementing the Bat Signal with your peers. When you're stuck and need a body double, a review, a brainstorm, an escape... this can be game-changing. Don't worry about being a bother, instead: pay it forward. We all need it a some point. 1/3
05.08.2025 00:00 — 👍 47 🔁 15 💬 1 📌 0
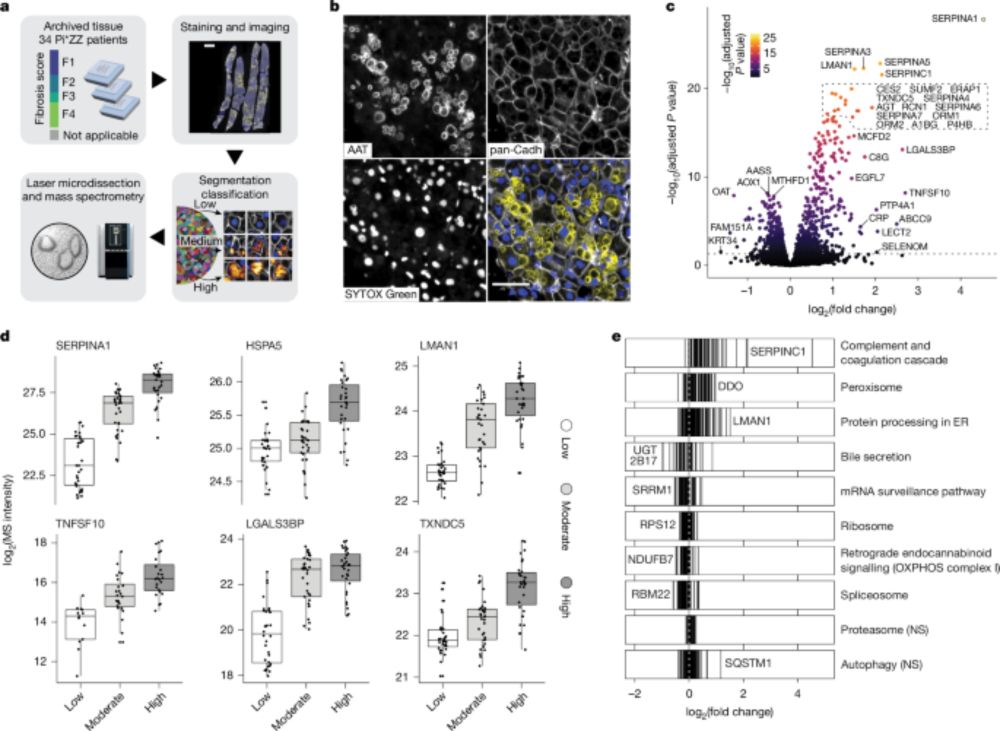
Conventional proteomics searches struggle with many modifications and fully open searches may be difficult to interpret. We introduce a "detailed" mass offset search in #MSFragger boosting interpretability and localization especially in complex cases (e.g. FPOP data): www.biorxiv.org/content/10.1...
01.08.2025 21:33 — 👍 34 🔁 6 💬 0 📌 0
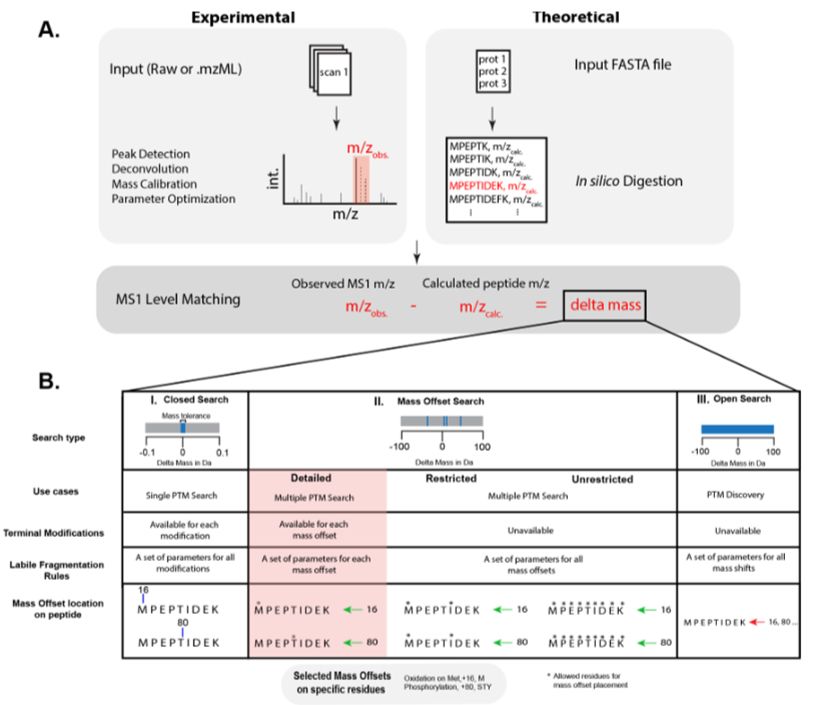
An automated workflow to address proteome complexity and the large search space problem in proteomics and HLA-I immunopeptidomics www.sciencedirect.co...
---
#proteomics #prot-paper
22.07.2025 11:20 — 👍 3 🔁 1 💬 0 📌 0
A perspective on integrating digital pathology, proteomics, clinical data and AI analytics in cancer research www.sciencedirect.co...
---
#proteomics #prot-paper
19.07.2025 11:20 — 👍 1 🔁 1 💬 0 📌 0
Some people seem to like the Bravo AssayMap, which has enrichment protocols using their proprietary cartridges and also autoSP3. There are several protocols using the Opentrons 2, but they aren't necessarily directly transferable compatible with the OT Flex, which also requires special tips.
27.06.2025 22:55 — 👍 1 🔁 0 💬 0 📌 0
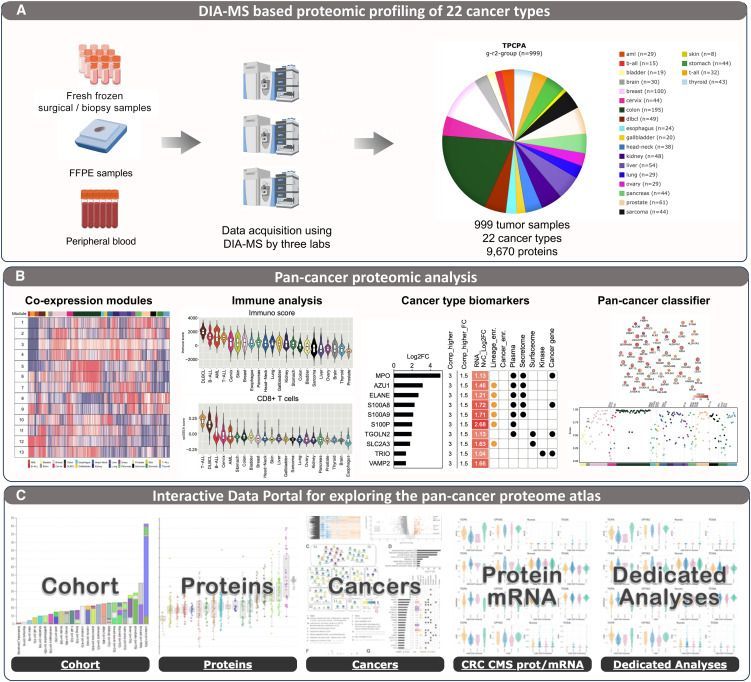
A unified pan-cancer proteome atlas www.cell.com/cancer-...
---
#proteomics #prot-paper
18.06.2025 20:00 — 👍 1 🔁 3 💬 0 📌 0
I had the opportunity to present MAETi, which enables #immunopeptidomics profiling of 10,000 cells. I was very happy to see that there is a lot of interest on it since @julbeyr.bsky.social , PhD student at Stefan Tenzer lab, has done a lot of great work optimizing it to reach these levels.
20.06.2025 16:45 — 👍 3 🔁 0 💬 0 📌 0
Megan Ford did an excellent job presenting our antibody-based workflow for high-throughput #immunopeptidomics of low-input cell and clinical samples. A big thanks to @dgpf.bsky.social sponsoring her travel award that made her assistance possible.
20.06.2025 16:45 — 👍 2 🔁 0 💬 1 📌 0
It felt like a big milestone to come back to France to present our work at the Immunopeptidomics Platform and Tenzer Lab HI-TRON Mainz, 7.5 years after my PhD in Paris:
20.06.2025 16:45 — 👍 0 🔁 0 💬 1 📌 0
Thanks a lot to the organizers and all the people who made it such a great experience. Merci beaucoup!
EuPA, FPS, and @ypic.bsky.social
20.06.2025 16:45 — 👍 1 🔁 0 💬 1 📌 0
#EuPA2025 was a blast !
Science was excellent and the personal discussions enriching. I was charmed to reconnect with the French proteomics community, and the breeze and beauty of Saint Malo made it unforgettable.
20.06.2025 16:45 — 👍 9 🔁 0 💬 2 📌 0
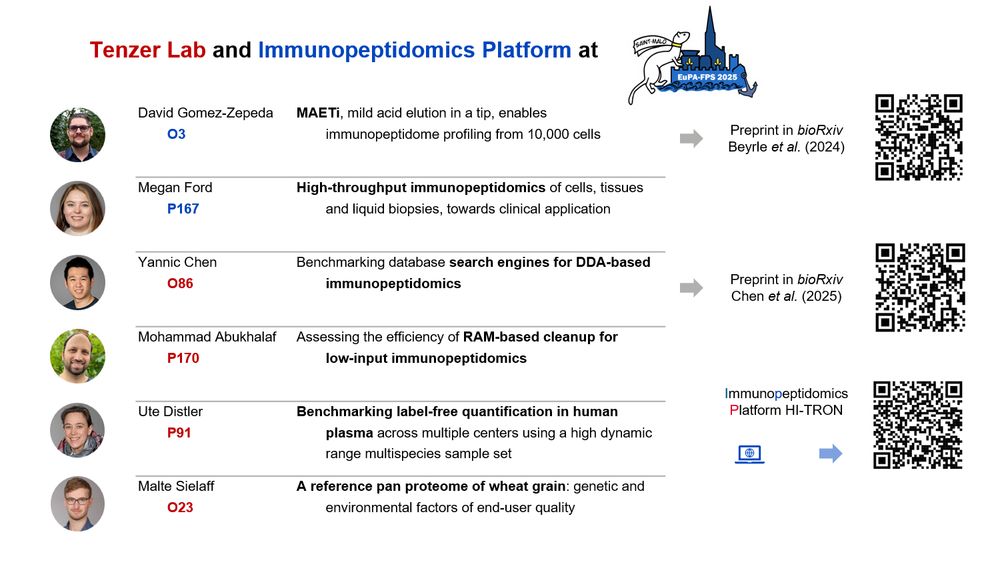
#TeamMassSpec attending the #EuPA2025 conference, don’t miss the presentations from the Tenzer Lab and Immunopeptidomics Platform to learn more about our recent work on #immunopeptidomics, and high-throughput and clinical #proteomics. See you in the beautiful Saint Malo!
16.06.2025 13:18 — 👍 18 🔁 3 💬 0 📌 0
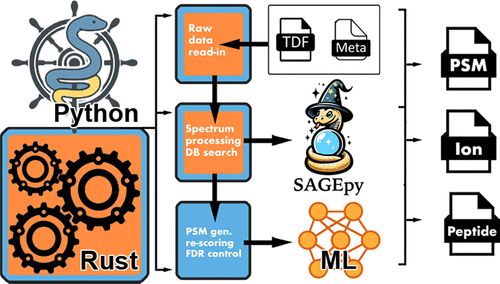
Rustims: An Open-Source Framework for Rapid Development and Processing of timsTOF Data-Dependent Acquisition Data pubs.acs.org/doi/10....
---
#proteomics #prot-paper
22.04.2025 15:40 — 👍 5 🔁 1 💬 0 📌 0
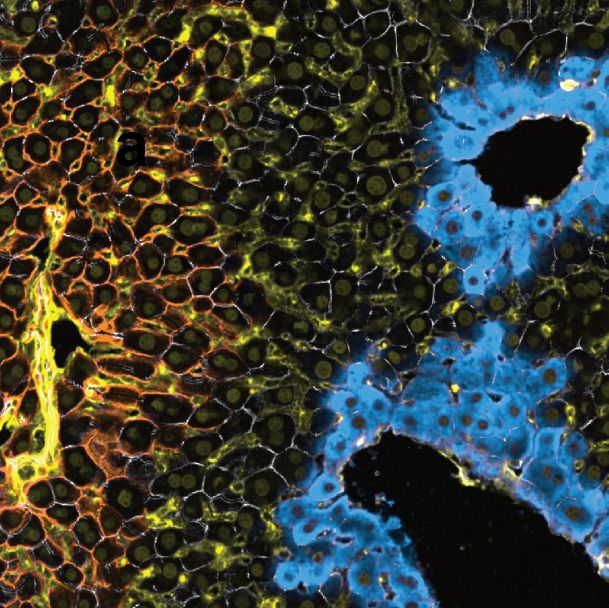
Immunostaining of liver tissue.
⏰ Don’t miss the chance to apply for this postdoc opportunity in spatial & single-cell proteomics in our lab at Karolinska Institutet @ki.se ! Plenty of exciting resources and projects on mitochondria and metabolism. Deadline: May 5 👉 bit.ly/4jXTxSK #mitochondria #metabolism #singlecell
30.04.2025 16:02 — 👍 5 🔁 2 💬 0 📌 0

The #Brixen Summer School on Proteomics is back! Join us for an immersive week of cutting-edge proteomics education and vibrant scientific exchange. To find the full program, follow this link: www.brixenproteomics.org/program/ . ❗Deadline for registration is 19th of May❗
21.04.2025 10:28 — 👍 7 🔁 5 💬 0 📌 1

Terminator holding a mass spectrometer saying "hasta la oxoprolina baby"
23.04.2025 16:32 — 👍 2 🔁 0 💬 0 📌 1

News in Proteomics Research blog post | Attention freeloaders! Skyline needs your support! It takes 60s to sign this thing! proteomicsnews.blogs...
---
#proteomics #prot-other
21.04.2025 22:40 — 👍 10 🔁 6 💬 0 📌 1
May Institute – Computation and statistics for mass spectrometry and proteomics
A few more spots are still open for some of the in-person programs at May Institute on Computation and Statistics for Mass Spectrometry and Proteomics on April 28 – May 11, 2025 on the campus of Northeastern University in Boston MA computationalproteomics.khoury.northeastern.edu
15.04.2025 18:10 — 👍 16 🔁 9 💬 0 📌 1
Join us! | Hoogendoorn Lab
Openings for PhD students and postdocs will be advertized here
Job alert: fully funded postdoc position in chemical biology in beautiful Geneva! Do you have a PhD in Chemical Biology or related field with organic synthetic experience? Apply now! Details on how to apply can be found here: www.hoogendoornlab.org/joinus #chemsky #ChemBio
03.12.2024 11:26 — 👍 25 🔁 23 💬 0 📌 2
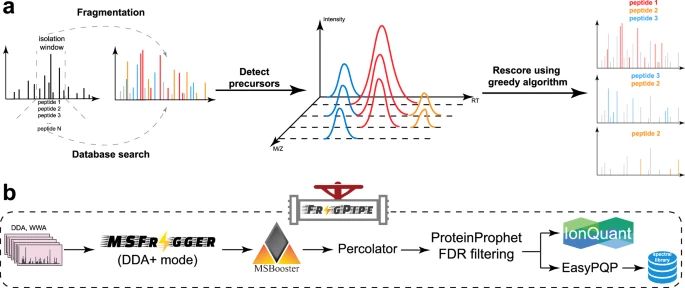
MSFragger-DDA+ enhances peptide identification sensitivity with full isolation window search www.nature.com/artic...
---
#proteomics #prot-paper
08.04.2025 18:00 — 👍 11 🔁 4 💬 0 📌 1
If you want to learn how to do #immunopeptidomics from <50,000 cells, join us at this Evosep webinar. @julbeyr.bsky.social will present Mild Acid Elution in a Tip (MAETi), a simple, antibody-free approach enabling unprecedented sensitivity
03.04.2025 14:43 — 👍 4 🔁 0 💬 0 📌 0
It depends on several factors: main application, budget, availability of service engineers in your region. Are your samples scarce? Do you aim for high-throughput? Mostly cell/tissue lysates or also plasma? Only proteomics or also peptidomics?
31.03.2025 23:01 — 👍 0 🔁 0 💬 0 📌 0

Rapid Determination of Isotopic Purity of Stable Isotope (D, 15N, or 13C)‐Labeled Organic Compounds by Electrospray Ionization‐High‐Resolution Mass Spectrometry
Rationale Stable isotope-labeled organic compounds, containing D, 15N, or 13C, have widespread applications in chemistry, biology, environmental science, and agriculture. However, the isotopic purit...
Rapid Determination of Isotopic Purity of Stable Isotope (D, 15N, or 13C)-Labeled Organic Compounds by Electrospray Ionization-High-Resolution Mass Spectrometry #RCM analyticalsciencejournals.onlinelibrary.wiley.com/doi/full/10....
26.03.2025 19:17 — 👍 1 🔁 1 💬 0 📌 0
If 21k UP is consensus while 63k UP includes isoforms, the redundancy in the latter may result in lower cut-off.
I guess that >100 AAs includes 21k UP while the others include 63k UP. If that's the case, I do see the upwards trend.
Note aside, I would plot cut-off (dependent variable) in y axis
21.03.2025 11:40 — 👍 2 🔁 0 💬 1 📌 0
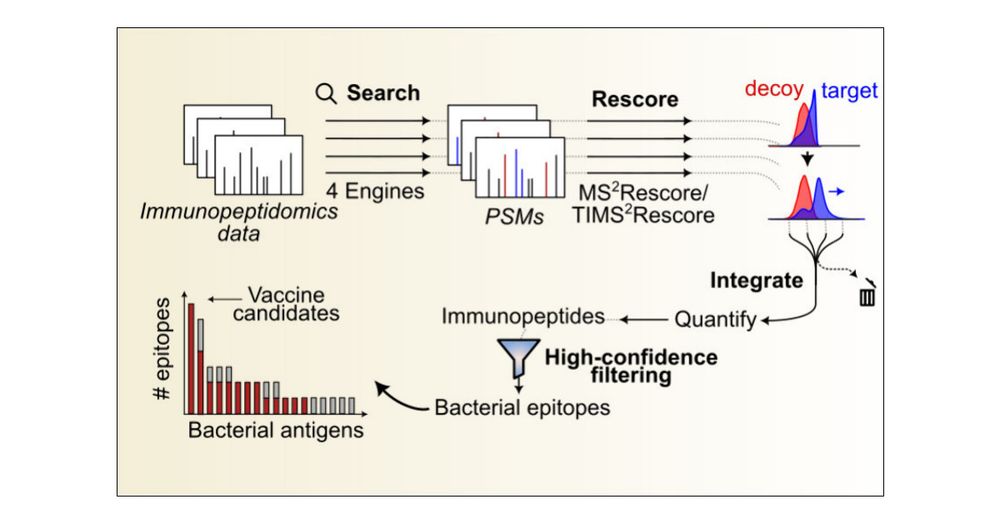
Maximizing Immunopeptidomics-Based Bacterial Epitope Discovery by Multiple Search Engines and Rescoring
Mass spectrometry-based discovery of bacterial immunopeptides presented by infected cells allows untargeted discovery of bacterial antigens that can serve as vaccine candidates. However, reliable identification of bacterial epitopes is challenged by their extremely low abundance. Here, we describe an optimized bioinformatic framework to enhance the confident identification of bacterial immunopeptides. Immunopeptidomics data of cell cultures infected with Listeria monocytogenes were searched by four different search engines, PEAKS, Comet, Sage and MSFragger, followed by data-driven rescoring with MS2Rescore. Compared with individual search engine results, this integrated workflow boosted immunopeptide identification by an average of 27% and led to the high-confidence detection of 18 additional bacterial peptides (+27%) matching 15 different Listeria proteins (+36%). Despite the strong agreement between the search engines, a small number of spectra (<1%) had ambiguous matches to multiple peptides and were excluded to ensure high-confidence identifications. Finally, we demonstrate our workflow with sensitive timsTOF SCP data acquisition and find that rescoring, now with inclusion of ion mobility features, identifies 76% more peptides compared to Q Exactive HF acquisition. Together, our results demonstrate how integration of multiple search engine results along with data-driven rescoring maximizes immunopeptide identification, boosting the detection of high-confidence bacterial epitopes for vaccine development.
🚀 Boosting bacterial immunopeptide discovery! Our bioinformatics workflow integrates 4 search engines with follow-up rescoring to enhance ID by 27%, maximizing the discovery of vaccine candidates. Applied to timsTOF/Thermo data🦠💉Great work by Patrick Willems.
pubs.acs.org/doi/10.1021/...
19.03.2025 20:17 — 👍 8 🔁 3 💬 0 📌 1
I took a very basic MS course during my studies. A lot of self learning from books and publications, and by doing it since I was the only one doing proteomics at the lab where I did my PhD. Two summer schools were essential:
EMBO Targeted Proteomics Barcelona and European Advanced Proteomics Brixen
20.03.2025 14:02 — 👍 3 🔁 0 💬 0 📌 0
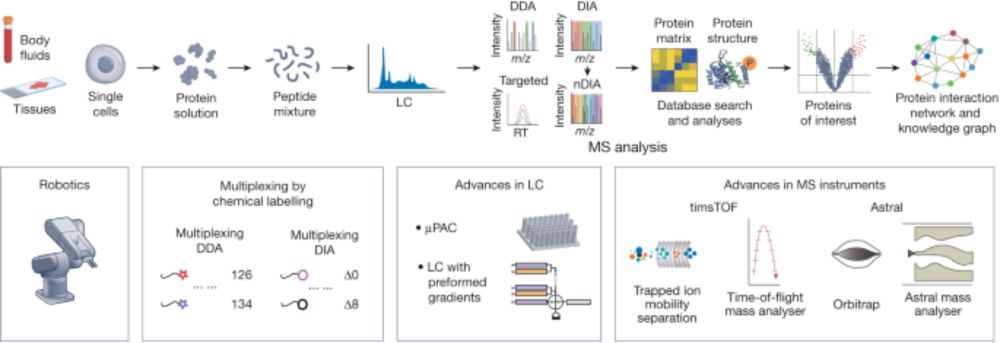
Mass-spectrometry-based proteomics: from single cells to clinical applications - Nature
This Review summarizes advances in mass-spectrometry-based proteomics and explores the potential applications of these technologies in the clinic.
In our latest @Nature review with Tiannan Guo & Judith Steen, we explore how technological breakthroughs are revolutionizing MS-based proteomics: From enhanced sensitivity enabling single-cell analysis to high-throughput plasma proteomics & AI-based data interpretation
www.nature.com/articles/s41...
26.02.2025 16:22 — 👍 14 🔁 9 💬 1 📌 0
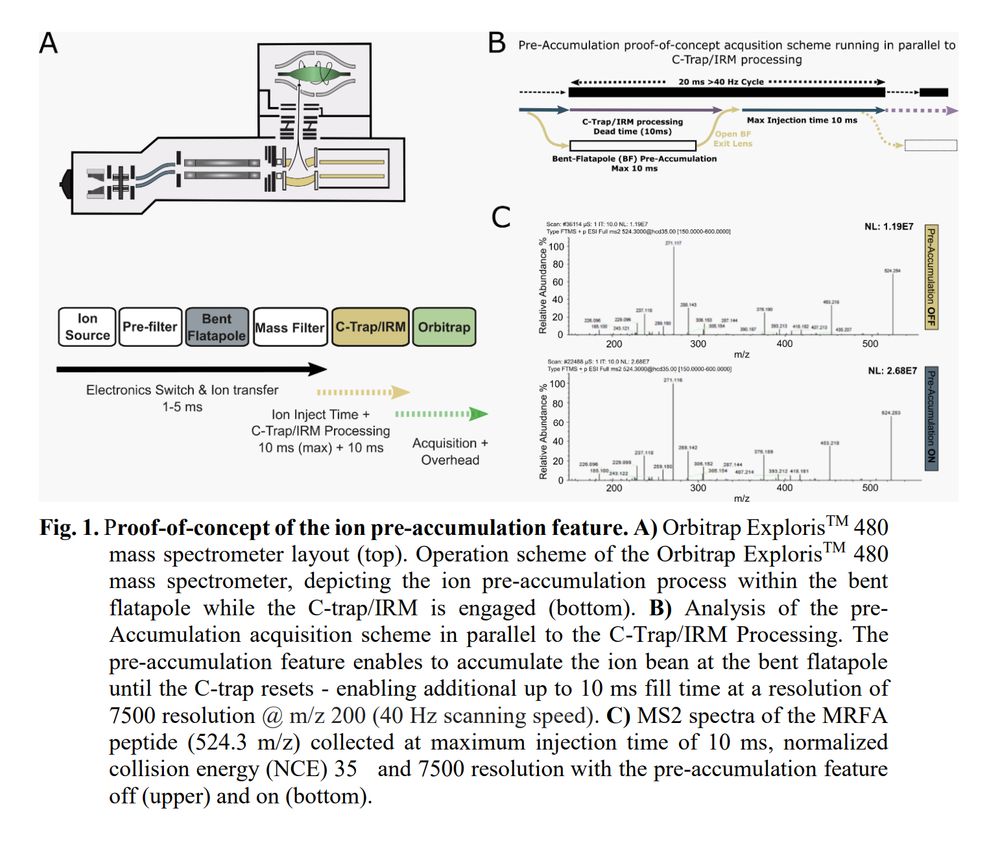
Enhancing tandem MS sensitivity and peptide identification via ion pre-accumulation in an Orbitrap mass spectrometer www.biorxiv.org/cont...
---
#proteomics #prot-preprint
26.02.2025 19:00 — 👍 10 🔁 2 💬 0 📌 1
Tenure-Track Professor in the field of Data Science in Mass Spectrometry-Based Omics
Tenure-Track Professor in the field of Data Science in Mass Spectrometry-Based Omics
We want you on our team!
If you’re an expert in computational mass spectrometry, machine-learning-driven omics & high-throughput molecule identification, apply now for our tenure-track position at @univie.ac.at
jobs.univie.ac.at/job/Tenure-T...
#Hiring #MassSpec #Bioinformatics #Jobs
25.02.2025 22:02 — 👍 4 🔁 3 💬 0 📌 0
Immunoinformatics at Mcgranahan lab @uclcancer Ex @eapm_bsc @bsc_cns @carrerasIJC - Stirring the pan at @eltallaret @embassat - he/him
Proteomes (ISSN 2227-7382) is an #openaccess, peer reviewed journal on all aspects of #Proteomics science, published quarterly online by @mdpiopenaccess.bsky.social
First AI Bioinformatician built for Proteomics and beyond. Fast, intuitive analysis for scientists. Built by top-tier former Google/Verily engineers. Check us out at www.tesorai.com.
Exploring Mass Spectrometry & the Origins of Life: Is Life a Common Cosmic Phenomenon?
Old person forgetting everything they ever knew about proteomics at speeds in excess of 270 Hz...
Syncell's Microscoop enables high-sensitivity spatial proteomics, allowing researchers to isolate and identify proteins at precise locations for discovery.
Delivering on the Promise of the Human Proteome
Senior Scientist at Evosep, focus on developing workflows for mass spectrometry based biolomecule characterization and proteome profiling. 2 wheels + an engine = fun!
Biological Mass Spectrometry enthusiast, trying to make sense of cellular hierarchies and heterogeneity
Extreme enzymes for #proteomics and specialty applications. One-step/5-minute digests w/ orthogonal coverage to trypsin; Bias for #membraneproteins. Link to Journal of Proteomics paper (https://c-bio.link/JPBJrW)
Berkeley, CA
CinderBio.com
Postdoc in computational proteomics @CompOmics @VIBLifeSciences - turning public data into tissue biomarkers with ML, AI, and a mission to make proteomics truly reusable
Postdoc and proteomics researcher at the University Medical Center Mainz.
LC-MS and proteomics enthusiast
Group Leader and Adjunct Professor 🧑🏫 | Public Health | Data Engineering & Visualization | 🧬 Bioinformatics & 📈 Mass Spec Enthusiast | 📍 Berlin | Passionate About Collaboration and Open Science 🔬✨
Developing cutting-edge analytical platforms for comprehensive studies of biological systems. Specializing in ambient ionization mass spectrometry profiling, imaging, and real-time volatile organic compounds (VOCs) detection.
Mass spec, proteomics, scientist, Ph.D., tennis player, Venus Williams super fan :-)
The home of premier fundamental discoveries, inventions and applications in the analytical and bioanalytical sciences.
Published by @rsc.org 🌐 Website: rsc.li/analyst-rsc