YouTube video by Human Pangenome Reference Consortium
The Human Pangenome Reference Consortium (HPRC) Q&A
This season, we’re grateful for the chance to share the work, voices, and perspectives of the HPRC community.
Our new Q&A video features members from across the consortium reflecting on the technologies, collaborations, and insights shaping the human pangenome.
Watch here: youtu.be/bJ7wkbwd5oU?...
24.11.2025 16:25 — 👍 2 🔁 1 💬 0 📌 0
Human Pangenome Reference Consortium
Human Pangenome Reference Consortium
Curious about the work behind the Human Pangenome Reference Consortium?
Discover how researchers worldwide are working together to build a more complete reference of human genetic variation.
Learn more at humanpangenome.org
05.11.2025 20:12 — 👍 1 🔁 0 💬 0 📌 0

Wishing everyone a spook-tacular Halloween! 👻
Whether you’re celebrating or just enjoying a well-earned weekend, we hope it’s a wickedly wonderful one!
31.10.2025 16:13 — 👍 1 🔁 0 💬 0 📌 0

We're live at #ASHG2025!
Don't forget to look out for HPRC member presentations, happening this week.
15.10.2025 16:16 — 👍 2 🔁 1 💬 0 📌 0

Next week, we're headed to #ASHG2025!
HPRC members will be sharing their latest research and insights throughout the week.
See who's presenting ⬇️
08.10.2025 19:02 — 👍 6 🔁 4 💬 1 📌 0
Human Pangenome Reference Consortium
Human Pangenome Reference Consortium
🌍 Researchers from around the world are joining forces through the Human Pangenome Reference Consortium to develop a more comprehensive and dynamic pangenome resource.
Learn more about our mission: humanpangenome.org
02.10.2025 16:20 — 👍 1 🔁 1 💬 0 📌 0

Human-specific gene expansions contribute to brain evolution
A complete human genome and functional modeling in zebrafish pinpoint gene expansions
that may underlie the evolutionary innovations of the human brain.
A complete telomere-to-telomere genome sequence reveals 213 human-specific gene families, including candidates shaping brain evolution.
Functional studies highlight roles for GPR89B and FRMPD2B in brain expansion and synapse signaling.
Read more in Cell:
www.cell.com/cell/abstrac...
26.09.2025 20:26 — 👍 5 🔁 1 💬 0 📌 0

HPRC will see you at #ASHG!
Join our workshop, The Human Pangenome: Data, Tools, and Workflows, on Oct 14 (10–12 pm) + look out for HPRC member presentations throughout the meeting.
Check out the presentations highlighted below, with more to come!
25.09.2025 20:15 — 👍 4 🔁 3 💬 0 📌 0

The Human Pangenome Reference Consortium is building a more complete and inclusive reference for human genetics, one that better reflects human variation and drives new discoveries.
Our story: humanpangenome.org
17.09.2025 19:09 — 👍 3 🔁 0 💬 0 📌 0
Partners
Partners
Collaboration drives progress.
From Coriell and the Genome Reference Consortium to Google and GA4GH, HPRC partners bring essential expertise to building the human pangenome.
Learn more about our partnerships:
humanpangenome.org/partners/
04.09.2025 13:42 — 👍 1 🔁 0 💬 0 📌 0

Exploring gene content with pangene graphs
AbstractMotivation. The gene content regulates the biology of an organism. It varies between species and between individuals of the same species. Although
Pangene, an innovative computational tool, maps gene orientation, order & copy-number changes across genomes. Applied to the human pangenome, it reveals both known variation & complex haplotypes.
Read more:
academic.oup.com/bioinformati...
29.08.2025 16:48 — 👍 3 🔁 1 💬 0 📌 0
HPRC Data Explorer
🔍 Explore the Human Pangenome!
Our interactive Data Explorer makes it easy to navigate sequencing, assemblies, annotations, and alignments from the HPRC.
Start exploring the data here:
data.humanpangenome.org/raw-sequenci...
27.08.2025 18:40 — 👍 2 🔁 2 💬 0 📌 0
65 Genomes Expand Our Picture Of Human Genetics
Researchers closely examined the genomes of 65 individuals to paint a more complex, and more complete, picture of human genetic diversity.
Tune in to @scifri.bsky.social as experts discuss the analysis of 65 genomes from individuals around the world and how this work advances our understanding of human genetics and genome function.
🎧 Listen here: www.sciencefriday.com/segments/65-...
13.08.2025 20:31 — 👍 2 🔁 1 💬 0 📌 0

🚌 School’s back in session, and like dedicated students everywhere, our ELSI team is working hard to explore the ethical, legal, and social implications of building a more comprehensive human reference genome.
Meet the team behind it: www.humanpangenome.org/elsi-team/
06.08.2025 15:18 — 👍 2 🔁 1 💬 0 📌 0
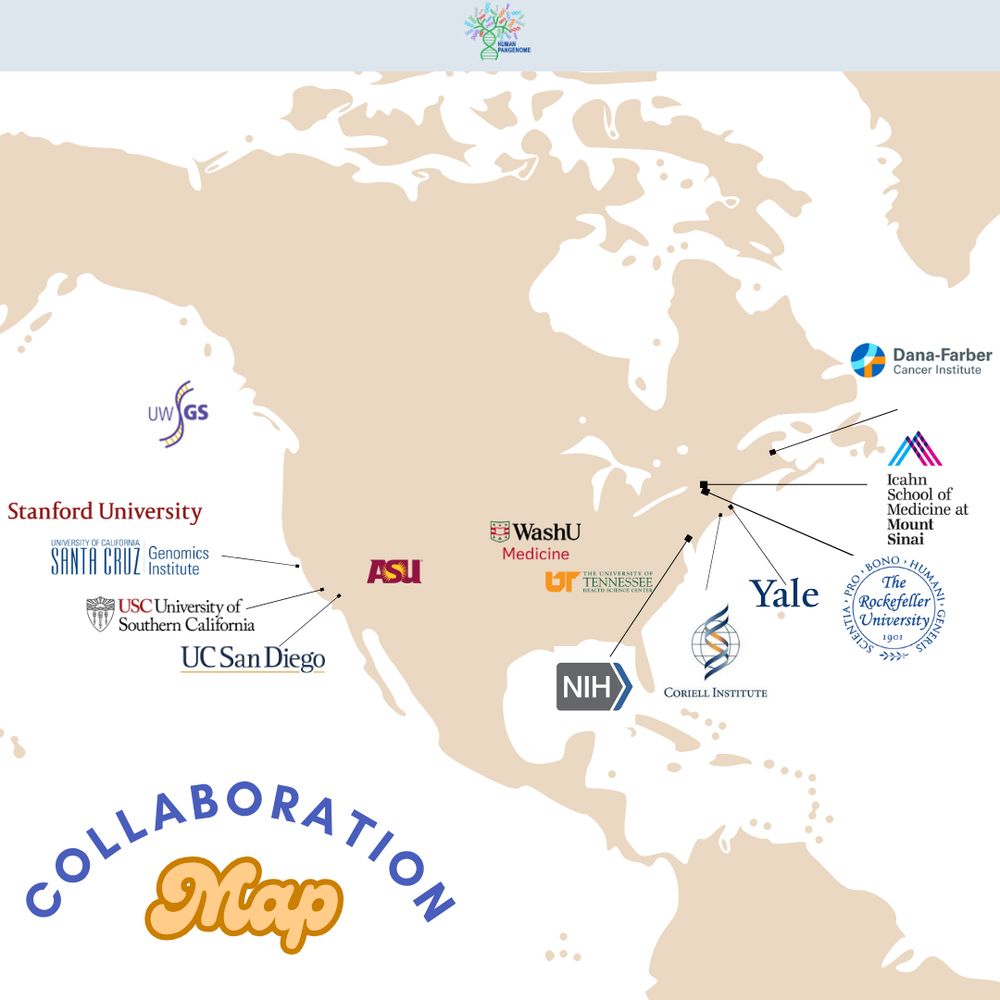
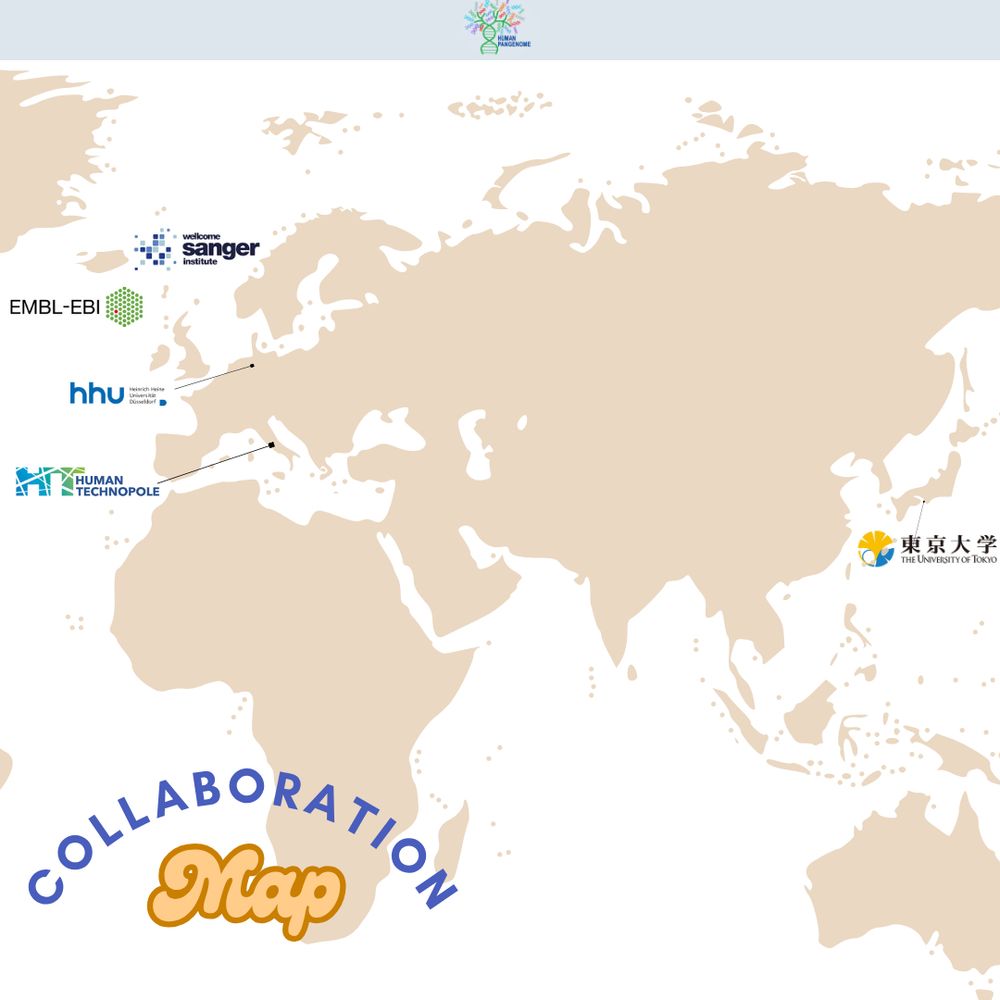
From the western hemisphere to the east 🌐 ... HPRC brings together institutions from across the globe.
Though separated by distance, we’re united by one common goal - building a pangenome reference that captures the full scope of human genomic variation.
18.07.2025 12:58 — 👍 1 🔁 1 💬 0 📌 0
Population Sampling and Representation
Population Sampling and Representation
A more complete understanding of human genetics starts with accurate, high-quality reference genomes.
HPRC is building a reference that reflects the breadth of human genetic variation, supporting better prediction, diagnosis, and treatment.
Learn more: humanpangenome.org/samples/
11.07.2025 20:38 — 👍 2 🔁 1 💬 0 📌 0
Highly accurate assembly polishing with DeepPolisher
An international, peer-reviewed genome sciences journal featuring outstanding original research that offers novel insights into the biology of all organisms
An emerging polishing pipeline, DeepPolisher, improves genome assemblies by reducing indel errors and correcting false homozygosity.
Applied to 180+ HPRC assemblies, it cut errors by 54% and significantly boosted overall quality.
Here’s how…
genome.cshlp.org/content/35/7...
09.07.2025 17:52 — 👍 2 🔁 0 💬 0 📌 0
PhD student at the NHGRI genomeinformatics.github.io & JHU Schatzlab
🛰️ 🧬🇨🇦
Where people matter, and serious work is done. Share your experiences with #WashU and look for @ WashU on Instagram and on TikTok. WashU.edu
CEO of Bluesky, steward of AT Protocol.
dec/acc 🌱 🪴 🌳
official Bluesky account (check username👆)
Bugs, feature requests, feedback: support@bsky.app
The Alliance of Genome Resources supports biological science via data integration, standardization of data models and interfaces, and unified outreach.
Find us at https://www.alliancegenome.org.
Scientist at Washington University in St Louis
How do you do, fellow squids? 🌊 We're your frondly Aquarium with a mission to inspire conservation of the ocean. https://www.montereybayaquarium.org
HealthDisparity Researcher who is decolonist, anticolonist, or recolonist/reconquistist. Whatever gets you to see that 1448-1492 was a bad road & that we have to start anew. The West is Dead, get over yourself. Pronouns are todos, omni, (A ∩ & ∪ Ω)
Oncologist-scientist at Weill Cornell and New York genome center. Into somatic evolution, trees and words
Computational Biologist. Mostly in English, sometimes in French. #NoPasaran
The world's largest open resource of human genetic variation. For help please use http://broad.io/gnomad_forum; feature requests/bug reports to http://broad.io/gnomad_github
Genomics plus anything else I find interesting. 🇨🇭🇵🇭 Opinions are my own and do not represent my employer.
I study cancer at Washington University in St Louis. Cancer Genomics, Bioinformatics, Data Viz, Tumor Evolution, AML, Immunotherapy, Irreverent humor 🧬 🖥️ mostly @chrisamiller on other platforms
Human geneticist turned data scientist in Seattle. Supporting clinicians at a large hospital. Human first. Kindness, standing up for people, sailing, climbing mountains, making a ruckus when necessary. Enduring optimist often tested to the limit.















